Volume 31, Number 9—September 2025
Research Letter
Monkeypox Virus Clade IIa Infections, Liberia, 2023–2024
Figure
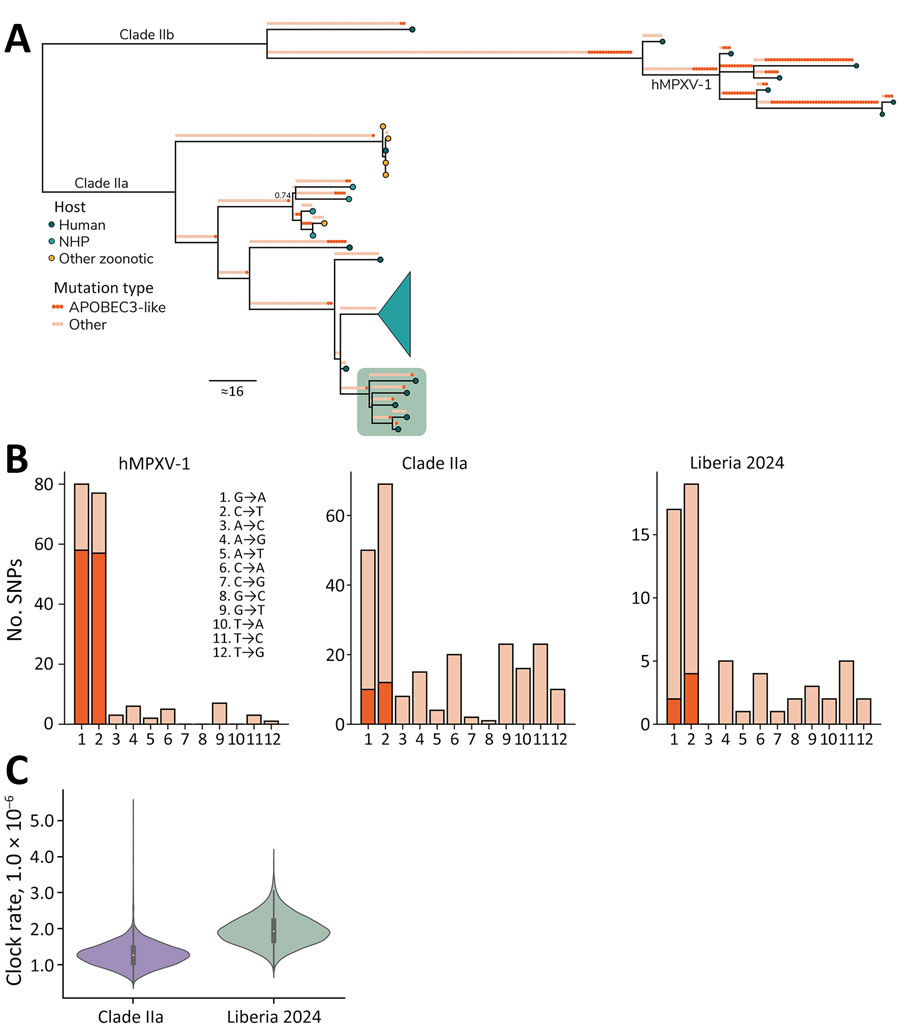
Figure. Sequencing characteristics of monkeypox virus clade IIa infections, Liberia, 2023–2024. A) Ancestral state reconstruction of the clade II phylogeny comparing the 5 new sequences against all publicly available clade IIa genomes (n = 27) and a subset of clade IIb sequences for context (n = 7), including hMPXV-1. Mutations are annotated along each branch with circles colored by whether it is an APOBEC3-like signature (TC→TT and GA→AA; dark orange) or whether it is another mutation type (light orange). Branch tips show circles colored by host (human, dark blue; NHP, turquoise; other, yellow). The sequences generated from this study are displayed in the green box. A large clade from Côte d'Ivoire NHPs is collapsed (turquoise funnel). Ultrafast bootstrap support is indicated only at nodes for which support <0.75. B) Number of APOBEC3-like SNPs of all mutations for subclade hMPXV-1, prior clade IIa sequences, and the new clade IIa found in Liberia. C) Comparison of the evolutionary clock rate of the prior clade IIa and new Liberian clade IIa sequences estimated under the local clock model. APOBEC3, apolipoprotein B mRNA editing enzyme catalytic subunit 3; hMPXV-1, sustained human monkeypox virus outbreak; NHP, nonhuman primate; SNP, single-nucleotide polymorphism.
1These first authors contributed equally to this article.