Volume 31, Number 9—September 2025
Dispatch
Related Melioidosis Cases with Unknown Exposure Source, Georgia, USA, 1983–2024
Figure 2
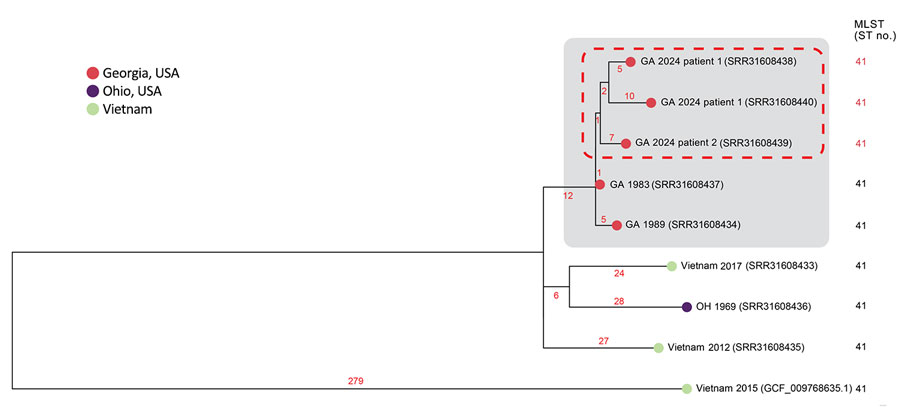
Figure 2. Subpanel maximum-likelihood phylogeny of refined mutation-only core single-nucleotide polymorphism sites of isolate genomes from 4 related melioidosis cases with unknown exposure source, Georgia, USA, 1983–2024, which includes the clinical isolates from the 2 patients from Georgia in 2024 (red dashed rectangle). B. pseudomallei genome sequences from the patient isolates were highly related to each other and to 2 other patients from Georgia in 1983 and 1989 (gray rectangle). Those genomes clustered most closely with genomes from Southeast Asia, particularly from Vietnam. Tree leaves contain year of isolation and National Center for Biotechnology Information sequence read archive and RefSeq accession numbers that correspond to each isolate. Branch numbers indicate the number of single nucleotide polymorphisms per site. MLST, multilocus sequence type; ST, sequence type.
1These first authors contributed equally to this article.