Volume 10, Number 11—November 2004
Dispatch
Burkholderia cenocepacia Vaginal Infection in Patient with Smoldering Myeloma and Chronic Hepatitis C
Cite This Article
Citation for Media
Abstract
We report a case of a vaginal infection caused by a strain of Burkholderia cenocepacia. The strain was isolated from vaginal swab specimens from a 68-year-old woman with smoldering myeloma and chronic hepatitis C virus infection who was hospitalized for abdominal abscess. Treatment with piperacillin/tazobactam eliminated B. cenocepacia infection and vaginal symptoms
Members of genus Burkholderia are aerobic, non–spore-forming, catalase-positive, gram-negative bacteria; most are oxidase positive (1). This genus comprises opportunistic pathogens responsible for important infections in immunocompromised persons and in cystic fibrosis (CF) patients (2,3). To date, the genus Burkholderia comprises more than 30 species, including the Burkholderia cepacia complex, B. mallei, and B. pseudomallei (2). The B. cepacia complex is a group of microorganisms composed of at least nine closely related genomovars (2,3). All genomovars have been shown to cause infections, and B. cenocepacia and B. multivorans (genomovars III and II, respectively) are the genomovars most frequently isolated from CF patients (4–7).
Nosocomial infections caused by B. cepacia complex have been reported in non-CF patients, principally associated with the use of contaminated disinfectants, anaesthetic solutions, and invasive treatments such as urinary and intravenous catheterization (8). These strains are intrinsically resistant to most antimicrobial agents and are difficult to eliminate (8,9). Cases of B. cepacia complex infections are underestimated because of the complex taxonomy of this genus and the poor sensitivity and specificity of commercial identification systems (10). Recently, molecular methods, mainly polymerase chain reaction (PCR)-based, have been developed to circumvent this issue (10–13).
We report a case of vaginal infection, caused by B. cenocepacia, in a patient affected by smoldering myeloma, and chronic hepatitis C virus (HCV) infection. Bacterial identification at species level was assessed by four combined polymerase-chain reaction (PCR)–based molecular methods. Therapy based on treatment with piperacillin/tazobactam completely eliminated the infection as well as the vaginal symptoms.
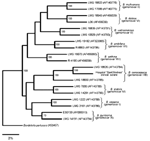
In August 2003, a 68-year-old woman with smoldering myeloma and chronic HCV infection (the patient had cirrhosis since 1994) was admitted to the “Sant’Andrea” Hospital (2nd Faculty of Medicine, “La Sapienza” University, Rome, Italy), with a 15-day history of fever, malaise, asthenia, fatigue, abdominal pain, and swellings from lower limbs. One week before admission, she had been treated with ciprofloxacin (500 mg twice a day) without improvement of any of the clinical symptoms. On admission (day 1), the patient had a fever (38.4°C) and showed abundant ascitic fluid and jaundice. Laboratory values were indicative of macrocytic anemia (erythrocytes, 3.6 x 109/L; hemoglobin, 110 g/L; mean corpuscular volume, 103 fL; hematocrit, 31%; serum iron level, 10.74 μmol/L; and serum ferritin level, 170 μg/L. Platelet count was 46 x 109/L, and leukocyte count was 14 x 109/L with neutrophils (13 x 109/L) and lymphocytes (0.5 x 109/L). The patient had high values of the erythrocyte sedimentation rate (ESR) in the first hour (71 mm/h) and C-reactive protein (CRP) (4.1 mg/L). Increased total serum proteins (8.1 g/dL) and hypoalbuminemia (23 g/L) were also detected. Six blood samples were taken at 3-hour intervals during the first day of hospitalization and cultured to detect the growth of aerobic and anaerobic microorganisms (Bactec System, Becton Dickinson, Sparks, MD). All blood cultures were negative. Abdominal ecographic and tomographic scans showed a pseudocystic formation in the pancreas. The pancreatic formation was drained because the patient was not suited for surgical intervention. On admission day 2, the patient was transferred to the Infectious Diseases Unit; there, intensive strong diuretic therapy was initiated, and a urinary catheter was inserted. Results of microbiologic analysis of urine and of a liquid taken from the pseudocystic formation were negative for common pathogenic bacteria. In spite of these results, the patient was given intravenous amoxicillin/clavulananate (1.2 g three times a day) and amikacin (1 g once a day) (day 3). After 5 days of antimicrobial drug therapy, the clinical symptoms of the patient slightly improved. On day 9 (i.e., after 6 days of antimicrobial drug therapy and 7 days of urinary catheterization), the patient exhibited an abundant white vaginal discharge with vulvar pain and burning. Vaginal swabs were streaked on different selective media. Columbia agar base, supplemented with 5% (vol/vol) sheep blood, and MacConkey agar plates showed a monomicrobic culture constituted by catalase-positive and oxidase-positive gram-negative rods that did not grow under anaerobic conditions. A presumptive identification of B. cepacia was made by using the API20NE (bioMérieux, Marcy l’Etoile, France), while the Vitek 2.0 identification system (bioMérieux) did not recognize the isolate as B. cepacia (10). Identification at the species level was achieved by four different PCR-based combined molecular methods, namely, DdeI and HaeIII restriction fragment length polymorphisms (RFLP) of 16S rDNA and recA gene analysis, recA genomovar-specific PCR, and recA sequence analysis (11–13). Control strains belonging to different genomovars of B. cepacia complex were included in all molecular analyses (14). Bacterial genomic DNA was extracted by using a commercial kit (Qiagen genomic-tip, Qiagen Inc., Hilden, Germany) as previously described (14). The bacterial isolate showed a 16S rRNA DdeI-RFLP pattern 1 and a recA HaeIII-RFLP pattern H (data not shown; 14), patterns indicative of B. cenocepacia (11–14). Genomovar-specific PCR was performed with primer pairs annealing to internal regions of the recA gene (12,13). A DNA fragment with a molecular mass of approximately 800 bp, consistent with the expected 781-bp B. cenocepacia amplification fragment, was successfully amplified with the primer pair BCRG3B1/BCRG3B2 (data not shown) (12,13). To unambiguously identify the bacterial isolate, we sequenced the amplified recA DNA fragment that was subjected to recA HaeIII-RFLP analysis (13). The recA DNA sequence of the bacterial isolate (GenBank accession no. AJ786367), subjected to BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST), showed >99% homology with the recA sequence of the B. cenocepacia reference strain LMG 18829 (GenBank accession no. AF143784). Phylogenetic analysis, based on recA DNA sequences, indicated that the clinical isolate belonged to the B. cenocepacia (genomovar III, lineage IIIB) (Figure) (4,13).
The B. cenocepacia isolate was resistant to penicillin, mezlocillin, piperacillin, amoxicillin/clavulanate, nitrofurantoin, ciprofloxacin, carbapenems, cephalosporins, aminoglycosides, and tetracycline and sensitive to trimethoprim/sulfamethoxazole and piperacillin/tazobactam. When the antimicrobial drug susceptibility profile was considered, the amoxicillin/clavulanate and amikacin antibiotic therapy was interrupted, and intravenous piperacillin/tazobactam combination was administered (4.5 g, three times a day for 4 weeks). Vaginal swabs were taken every 3 days during the 4 weeks of the antimicrobial drug therapy and, afterwards, every 20 days for a total follow-up period of 3 months. After 10 days of the piperacillin/tazobactam treatment, vaginal symptoms disappeared and cultured vaginal swabs did not show B. cenocepacia. After the piperacillin/tazobactam treatment ended, the patient did not exhibit any signs of vaginal infection.
We think this is the first description of a vaginal infection caused by B. cenocepacia. The patient’s immunodepression from smoldering myeloma and chronic HCV likely favored vaginal colonization by B. cenocepacia. Urinary catheterization might have favored vaginal colonization by B. cenocepacia, even if we did not isolate B. cenocepacia from catheters, disinfectants, and selected hospital environmental samples analyzed from October 2003 to date February 2004 (15). Moreover, the antimicrobial agents, ciprofloxacin, and amoxicillin/clavulanate and amikacin, administered to the patient before and during hospitalization, might also have altered the patient’s vaginal flora. Piperacillin/tazobactam eliminated vaginal symptoms and B. cenocepacia from the vaginal mucosa, thus indicating that the detected isolate was indeed responsible for the infection.
Microorganisms belonging to the B. cepacia complex are difficult to identify by conventional biochemical tests and commercial systems (8). This case report highlights the importance of the use of molecular techniques to quickly and accurately identify members of the B. cepacia complex (10,13). The ability of B. cenocepacia to cause vaginal infections is unusual. Further studies are needed to clarify whether specific virulence factors are carried and expressed by the B. cenocepacia clinical isolate, conferring to this strain the specific ability to colonize and multiply within the vaginal mucosa.
Mr. Petrucca is a medical biotechnology student who is doing research at the “Sant’Andrea” Hospital, 2nd Faculty of Medicine, “La Sapienza” University, Rome, Italy. His main research interests include developing new tools to identify bacterial pathogens and regulate gene expression of type III secretion systems in enteroinvasive bacteria.
Acknowledgments
We thank Styliani Papadopoulou and Cristina Petrucci for their excellent technical assistance.
This work was supported by faculty 60% funds granted to R.S. and to M.N., and in part by MURST PRIN research project “Effettori di virulenza in patogeni enterici: caratteristiche e studio delle loro interazioni,” granted to M.N.
References
- Gillan PH, Whittier S. Burkholderia, Stenotrophomonas, Ralstonia, Brevundimonas, Comamonas and Acidovorax In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolker RH, editors. Manual of clinical microbiology. Washington: American Society of Microbiolgy; 1999. p. 526–38.
- Coenye T, Vandamme P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol. 2003;5:719–29. DOIPubMedGoogle Scholar
- Coenye T. LiPuma JJ. Molecular epidemiology of Burkholderia species. Front Biosci. 2003;8:e55–67. DOIPubMedGoogle Scholar
- Vandamme P, Holmes B, Coenye T, Goris J, Mahenthiralingam E. LiPuma JJ, Govan JRW. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res Microbiol. 2003;154:91–6. DOIPubMedGoogle Scholar
- Speert DP, Henry DA, Vandamme P, Corey M, Mahenthiralingam E. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg Infect Dis. 2002;8:181–7.PubMedGoogle Scholar
- Agodi A, Mahenthiralingam E, Bachitta M, Giannino V, Sciacca A, Stefani S. Burkholderia cepacia complex infection in Italian patients with cystic fibrosis: prevalence, epidemiology, and genomovar status. J Clin Microbiol. 2001;39:2891–6. DOIPubMedGoogle Scholar
- LiPuma JJ. Spilker T, Gill GH, Campbell PW, Liu L, Mahenthiralingam E. Disproportionate distribuition of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am J Respir Crit Care Med. 2001;164:92–6.PubMedGoogle Scholar
- Coenye T, Vandamme P, Govan JRW. LiPuma JJ. Taxonomy and identification of the Burkholderia cepacia complex. J Clin Microbiol. 2001;39:3427–36. DOIPubMedGoogle Scholar
- Nzula S, Vandamme P, Govan JR. Influence of taxonomic status on the in vitro antimicrobial susceptibility of the Burkholderia cepacia complex. J Antimicrob Chemother. 2002;50:265–9. DOIPubMedGoogle Scholar
- Henry DA, Mahenthiralingam E, Vandamme P, Coenye T, Speert DP. Phenotypic methods for determining genomovar status of the Burkholderia cepacia complex. J Clin Microbiol. 2001;39:1073–8. DOIPubMedGoogle Scholar
- Vandamme P, Henry D, Coenye T, Nzula S, Vancanneyt M, LiPuma JJ, et al. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol Med Microbiol. 2002;33:143–9. DOIPubMedGoogle Scholar
- Vermis K, Coenye T, Mahenthiralingam E, Nelis HJ, Vandamme P. Evaluation of species-specific recA-based PCR tests for genomovar level identification within the Burkholderia cepacia complex. J Med Microbiol. 2002;51:937–40.PubMedGoogle Scholar
- Mahenthiralingam E, Bischof J, Byrne SK, Radomski C, Davies JE, Av-Gay Y, DNA-based diagnostic approaches for the identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, Burkholderia cepacia genomovars I and III. J Clin Microbiol. 2000;38:3165–73.PubMedGoogle Scholar
- Petrucca A, Cipriani P, Valenti P, Santapaola D, Cimmino C, Scoarughi GL, Molecular characterization of Burkholderia cepacia isolates from cystic fibrosis (CF) patients in an Italian CF center. Res Microbiol. 2003;154:491–8. DOIPubMedGoogle Scholar
- Siddiqui AH, Mullingam ME, Mahenthiralingam E, Hebden J, Brewrink J, Qaiyumi S, An episodic outbreak of genetically related Burkholderia cepacia among non–cystic fibrosis patients at a university hospital. Infect Control Hosp Epidemiol. 2001;22:419–22. DOIPubMedGoogle Scholar
Figure
Cite This ArticleTable of Contents – Volume 10, Number 11—November 2004
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Mauro Nicoletti, Dipartimento di Scienze Biomediche, Sezione di Microbiologia–Università “G. D’Annunzio,” Via dei Vestini 31, 66100, Chieti, Italy; fax: +39-06-49914626
Top