Volume 12, Number 9—September 2006
Research
Genomic Signatures of Human versus Avian Influenza A Viruses
Figure
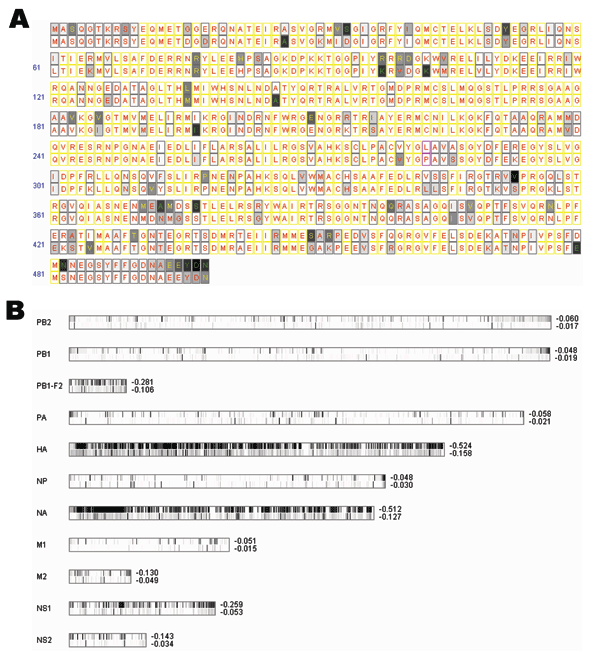
Figure. A) Entropy plot for avian versus human influenza viruses for NP amino acid residues. In each aligned position, we have a consensus residue for 95 avian strains displayed on top and a consensus residue for 306 human strains at the bottom. Completely conserved amino acid positions are filled with white; less conserved amino acids are filled in various gray shadings. Positions in which 1 single residue dominates >90%, <90% but >75%, and <75% are labeled with red, yellow, and green letters, respectively. Yellow rectangles indicate that both human and avian viruses are completely conserved to the same residue; magenta rectangles indicate that avian and human viruses are each completely conserved to a different residue. B) Entropy plots for the entire influenza A viral genome. Each lane displays entropy value distributions of aligned protein sequences for 1 of the 11 viral proteins; the upper half represents 95 avian strains, and the bottom half represents 306 human strains. (PB1-F2 contains fewer strains, as described in Discussion.) Positions completely conserved to a single residue are shown in a white band, while less conserved ones are shown in various gray shadings. The average entropy for the entire segment is shown to the right of these lanes. Entropy values are zero when residues are completely conserved; more negative values indicate more diversity. Alignment size for each protein from top to bottom is 759, 757, 90, 716, 591, 498, 480, 252, 97, 230, and 121.
1These authors contributed equally to this article.