Volume 14, Number 10—October 2008
Dispatch
Molecular Surveillance for Multidrug-Resistant Plasmodium falciparum, Cambodia
Cite This Article
Citation for Media
Abstract
We conducted surveillance for multidrug-resistant Plasmodium falciparum in Cambodia during 2004–2006 by assessing molecular changes in pfmdr1. The high prevalence of isolates with multiple pfmdr1 copies found in western Cambodia near the Thai border, where artesunate–mefloquine therapy failures occur, contrasts with isolates from eastern Cambodia, where this combination therapy remains highly effective.
The Thailand–Cambodia border has been an epicenter for drug-resistant Plasmodium falciparum. Recent clinical studies indicate that efficacy of artesunate–mefloquine combination is decreasing on both sides of the border (1–3). In contrast, P. falciparum in eastern Cambodia remains sensitive to mefloquine and the artesunate–mefloquine combination (4,5). Declining artesunate–mefloquine efficacy on the Thai border and the geographic variation in susceptibility to this combination therapy suggest expanded surveillance is needed in Cambodia. An inexpensive method to help target in vivo monitoring sites is surveillance for molecular markers of drug resistance.
We have previously shown that elevated P. falciparum multidrug resistance 1 (pfmdr1) gene copy number is associated with an 8-fold risk for artesunate–mefloquine failure in western Cambodia (6). More recently, we conducted a molecular surveillance for pfmdr1 mutations to guide the selection of sentinel sites for in vivo monitoring of artesunate–mefloquine resistance.
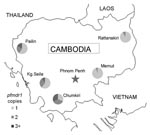
Clinical isolates of P. falciparum were collected from 5 sites across Cambodia (Figure 1): Pailin, Kampong Seila, Chumkiri (western), Memut, and Rattanakiri (eastern) during 2004–2006. Study participants included patients who were seen at health centers with uncomplicated falciparum malaria, including mixed infections. Fifty-six samples from patients in Pailin had been previously analyzed (6). Institutional review board approvals were obtained from the Cambodian National Ethics Committee for Health Research, the US Naval Medical Research Unit No. 2, and the University of North Carolina at Chapel Hill (UNC).
Peripheral blood was used to prepare smears and filter paper blood spots (25 μL, 2 spots) on Whatman 3-mm blotting paper (Whatman International Ltd., Maidstone, Kent, UK) from each participant. Blood smears were examined to determine parasite density, nonfalciparum species, and gametocytes. Enrolled participants completed a questionnaire on demographic information and past medical history. They received artesunate–mefloquine in accordance with national guidelines for uncomplicated falciparum malaria. DNA was extracted from blood spots by using QIAmp Mini kits (QIAGEN, Valencia, CA, USA). pfmdr1 copy number and single-nucleotide polymorphism assays were performed as previously described (6,7), except that in pfmdr1 single-nucleotide polymorphism assays the concentration of probes was reduced from 250 nM to 125 nM and reactions were not duplicated.
Linear regression was used to determine if pfmdr1 copy number varied by site. pfmdr1 copy number was inverse transformed to meet assumptions of normality and homoscedasticity. Backwards elimination based on a p value of 0.05 was used to determine the effect of site when controlling for confounders. Logistical regression was used to confirm the results of the linear regression analysis by using a cutoff of 1.5 copies (8). Logistic regression was also used to determine if pfmdr1-184 genotype varied by site. Samples with a mixture of 184-phe and 184-tyr were coded as 184-phe. All statistical analyses were conducted in Stata 8.2 (StataCorp, College Station, TX, USA).
We enrolled 744 study participants with uncomplicated P. falciparum malaria. The characteristics of participants at each site are described in Table 1. pfmdr1 copy number was successfully determined for 712 (95.7%) of 744 samples. When compared with results at UNC, results of copy number assays performed by the National Malaria Control Program (NMCP) staff in Phnom Penh were slightly lower (mean difference –0.163, 95% confidence interval [CI] –0.277 to –0.049, n = 44), and the 2 groups were in 100% concordance with distinguishing samples with >2 pfmdr1 copies.
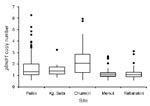
pfmdr1 copy numbers ranged from 0.6 to 6.3, and 167 (23.5%) of 712 samples had >1.5 copies. The median and distribution of pfmdr1 copy number varied by site and were highest in the 3 western Cambodian sites (Figure 2). The proportion of samples with >1.5 pfmdr1 copies was greater in western than eastern Cambodia (52.7% vs. 6.4%, odds ratio [OR] = 16.2, 95% CI 10.3–25.3). The prevalence of parasite samples with amplified pfmdr1 varied by site and differed in the prevalence of 2 and >3 copies of pfmdr1 (Figure 1).
The prevalence of pfmdr1-184-Phe varied by site, was significantly higher in the West (85.5% vs. 17.1%, OR = 27.4, 95% CI 18.1–41.4), but was not more prevalent in samples with pfmdr1 copy number >1.5 when site was controlled for (OR = 1.5, 95% CI 0.9–28, p = 0.139). Pfmdr1 codons 86, 1034, and 1042 were predominantly wild-type, 356 (98.6%) of 361, 215 (97.7%) of 220, 347 (95.9%) of 362, respectively, with little variation between sites (data not shown). Most (196 [91.2%] of 215) samples had the haplotypes 86-Asn, 1034-Ser, and 1042-Asn.
In univariate and multivariate linear regression models, site was the strongest predictor of pfmdr1 copy number. Chumkiri had the highest pfmdr1 copy number before and after clinical correlates were controlled for (Table 2). The relationship between copy number and site was retained when the analysis was repeated with logistic regression. Compared with Rattanakiri, the OR of having a copy number >1.5 was 32.4 (95% CI 16.4–64.2) for Chumkiri, 7.8 (95% CI 4.3–13.9) for Pailin, and 0.5 (95% CI 0.2–1.2) for Memut when parasitemia and year were controlled for.
We found geographic heterogeneity for 2 pfmdr1 genetic markers: pfmdr1 184 and pfmdr1 amplification. Genotype 184-Phe was associated with pfmdr1 amplification at the population level but not in individual isolates. The basis of this trend is unknown. Pailin, Kampong Seila, and Chumkiri in western Cambodia had higher pfmdr1 copy numbers and higher prevalence of pfmdr1 184-Phe than Memut and Rattanakiri in eastern Cambodia. An association between these markers and resistance is consistent with reports of reduced efficacy of artesunate–mefloquine in Pailin (79% over 42 days) (2) compared with Memut and Rattanakiri, where artesunate–mefloquine efficacy remains close to 100% (4,5). This finding is also consistent with the decrease of mefloquine sensitivity in western Cambodia based on in vitro drug susceptibility monitoring conducted by the Pasteur Institute of Cambodia from 2001 through 2007 (9,10; P. Lim, pers. comm.).
Chumkiri was surveyed because local health staff observed that falciparum malaria patients treated with artesunate–mefloquine often returned within weeks with recurrent fever and parasitemia. The possibility of such resistance in this site was particularly alarming because it was thought to be confined to Thailand–Cambodia border areas. Chumkiri had not previously been a sentinel site for antimalarial efficacy monitoring by NMCP. On the basis of these pfmdr1 data, a clinical trial of artesunate–mefloquine was launched in Chumkiri in 2006 (11). Clinical validation of predicted resistance by in vivo studies is beyond the scope of this study, although multiple in vitro and in vivo studies have consistently shown an association of increased pfmdr1 copy number with mefloquine or artesunate–mefloquine failure in Southeast Asia (8,10,12–15), supporting use of pfmdr1 copy number in routine surveillance and policy formation.
Monitoring changes in antimalarial drug efficacies is essential for guiding treatment policies in an era of multidrug resistance. However, such studies are resource intensive. In Cambodia, for example, NMCP can conduct in vivo studies at only 2–3 sites per year because of limited funds and trained staff. Molecular markers can help target in vivo studies where they are most needed. Molecular surveillance is high-throughput and can be performed in a central laboratory on dried blood spots. Expanded surveillance allows for molecular mapping and enables a rapid containment of resistance foci. pfmdr1 assays are now routinely performed in Cambodia by NMCP staff. National and regional molecular surveillance by malaria-endemic countries is a real possibility.
Mr Shah is in training to be a physician and infectious disease epidemiologist in training in the MD/PhD program at the University of North Carolina. His research interests include the epidemiology and molecular mechanisms of antimalarial resistance.
Acknowledgments
We thank the staff of Memut Referral Hospital, Rattanakiri Provincial Health Department, Pailin Referral Hospital, and Trapeang Raeng Health Center in Kampong Seila for their field assistance and support. We are also grateful to Anne Purfield and Parath Lim for laboratory assistance and to J. Kevin Baird and William O. Rogers for their encouragement and administrative support. MR4 provided control DNA samples of P. falciparum strains 3D7 and Dd2.
This work was funded by the US Navy Medical In-House Laboratory Independent Research Program at Naval Health Research Center/Naval Medical Research Center, Department of Defense Global Emerging Infections Systems, International Atomic Energy Agency (CRP E1.50.19), and the National Institutes of Health’s Fogarty International Center (PD 5D43TW007402-02).
References
- Vijaykadga S, Rojanawatsirivej C, Cholpol S, Phoungmanee D, Nakavej A, Wongsrichanalai C. In vivo sensitivity monitoring of mefloquine monotherapy and artesunate-mefloquine combinations for the treatment of uncomplicated falciparum malaria in Thailand in 2003. Trop Med Int Health. 2006;11:211–9. DOIPubMedGoogle Scholar
- Denis MB, Tsuyuoka R, Poravuth Y, Narann TS, Seila S, Lim C, Surveillance of the efficacy of artesunate and mefloquine combination for the treatment of uncomplicated falciparum malaria in Cambodia. Trop Med Int Health. 2006;11:1360–6. DOIPubMedGoogle Scholar
- Wongsrichanalai C, Meshnick SR. Declining artesunate–mefloquine efficacy against falciparum malaria on the Cambodia–Thailand border. Emerg Infect Dis. 2008;14(5):716–19 [cited 2008 Aug 6]. Available from http://www.cdc.gov/EID/content/14/5/716.htm
- Annual progress report of the National Center for Parasitology, Entomology and Malaria, 2004. Phnom Penh (Cambodia): The Center; 2005.
- Annual progress report of the National Center for Parasitology, Entomology and Malaria, 2006. Phnom Penh (Cambodia): The Center; 2007.
- Alker AP, Lim P, Sem R, Shah NK, Yi P, Bouth DM, pfmdr1 genotype and in vivo resistance to mefloquine-artesunate in falciparum malaria on the Thai-Cambodian border. Am J Trop Med Hyg. 2007;76:641–7.PubMedGoogle Scholar
- Purfield A, Nelson A, Laoboonchai A, Congpuong K, McDaniel P, Miller RS, A new method for detection of pfmdr1 mutations in Plasmodium falciparum DNA using real-time PCR. Malar J. 2004;3:9. DOIPubMedGoogle Scholar
- Price RN, Cassar C, Brockman A, Duraisingh M, van Vugt M, White NJ, The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob Agents Chemother. 1999;43:2943–9.PubMedGoogle Scholar
- Lim P, Chim P, Sem R, Nemh S, Poravuth Y, Lim C, In vitro monitoring of Plasmodium falciparum susceptibility to artesunate, mefloquine, quinine and chloroquine in Cambodia: 2001–2002. Acta Trop. 2005;93:31–40. DOIPubMedGoogle Scholar
- Containment of malaria multi-drug resistance on the Cambodia-Thailand Border Report of an Informal Consultation/Phnom Penh. 2007 Jan 29–30. SEA-MAL-246. WHO/SEARO, WHO/WPRO [cited 2008 Aug 6]. Available from www.who.int/malaria/docs/drugresistance/ReportThaiCam.pdf
- Sem R, Lim P, Muth S, Kim S, Chim P, Duong S, Efficacy of current standard therapy for uncomplicated P. falciparum and P. vivax malaria in central Cambodia. Abstracts of the Joint International Tropical Medicine Meeting 2007 “Health Security in the Tropics,” 2007 Nov 29–30; Bangkok. Abstract No. 47.
- Nelson AL, Purfield A, McDaniel P, Uthaimongkol N, Buathong N, Sriwichai S, pfmdr1 genotyping and in vivo mefloquine resistance on the Thai-Myanmar border. Am J Trop Med Hyg. 2005;72:586–92.PubMedGoogle Scholar
- Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, Zalewski C, Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47:2418–23. DOIPubMedGoogle Scholar
- Wilson CM, Volkman SK, Thaithong S, Martin RK, Kyle DE, Milhous WK, Amplification of pfmdr 1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol Biochem Parasitol. 1993;57:151–60. DOIPubMedGoogle Scholar
- Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 14, Number 10—October 2008
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Chansuda Wongsrichanalai, Infectious Diseases Team, Office of Public Health, USAID/Regional Development Mission – Asia (RDM-A), GPF Witthayu Tower A, 3rd Floor, 93/1 Wireless Rd, Bangkok 10330, Thailand;
Top