Volume 14, Number 11—November 2008
Dispatch
Phylogenetics and Pathogenesis of Early Avian Influenza Viruses (H5N1), Nigeria
Cite This Article
Citation for Media
Abstract
Three highly pathogenic avian influenza subtype H5N1 and 4 Newcastle disease viruses were isolated from sick or dead chickens in southwestern Nigeria. Sequencing and phylogenetic analysis placed them within H5N1 subclade 2.2.2. Intravenous and intranasal pathogenicity tests produced systemic disease with vascular endothelial cell tropism in chickens.
The first official report of avian influenza virus (H5N1) in Africa was made in January 2006 (1). Before then, surveillance was ongoing at the poultry clinic of the University of Ibadan Veterinary Teaching Hospital to identify causes of death in chickens in Nigeria.
Nasopharyngeal and cloacal swab samples were collected from sick and dead birds found on farms near Ibadan, Nigeria, and injected into 10-day-old embryonating chicken eggs. Retrospective analysis of isolates obtained in or near January 2006 identified influenza virus (H5N1) in 3 samples. In addition, 4 Newcastle disease virus isolates were obtained, which highlights the cocirculation of Newcastle disease virus and influenza viruses (H5N1) in Nigerian poultry and emphasizes the need for specific virologic testing to distinguish the clinically similar poultry diseases caused by these 2 pathogens.
The 3 avian influenza (H5) isolates were designated influenza A/chicken/Nigeria/228-5/2005, A/chicken/Nigeria/228-6/2006, and A/chicken/Nigeria/228-10/2006. Nucleotide sequences of the coding regions of all 8 segments of the 3 viruses demonstrated that all isolates possessed a multiple basic amino acid at the hemagglutinin (HA) cleavage site with the sequence PQGERRRKKR. Sequences at this site were identical to those of highly pathogenic avian influenza (HPAI) subtype H5N1 viruses from Europe, Russia, Asia, and from recent isolates from the Lagos state of Nigeria (2); it lacks a single basic residue when compared with HA from strains from southeastern People’s Republic of China, Vietnam, Cambodia (PQRERRRKKRG), and Thailand (PQREKRRKKRG) (3). Other notable features of the sequences were the absence of the H274Y genetic change associated with high-level resistance to oseltamivir in influenza neuraminidase 1 (N1) viruses (4). Similarly, known amantadine resistance–linked mutations were absent. The nonstructural (NS) 1 open reading frame encodes a 5-aa deletion at positions 80–84, as has been observed since 2001 in subtype H5N1 isolates from poultry. One of the isolates (A/chicken/Nigeria/228-10/2006) also has a C-terminal amino-acid extension, which is predicted to affect the function of the PDZ-ligand domain otherwise present at the C terminus of the NS1 protein (5,6). This sequence change did not detectably affect the ability of NS1 to block interferon induction when expressed transiently in 293T cells (data not shown). The polymerase basic 2 protein (PB2) of these viruses possesses a lysine residue at position 627, an amino acid previously implicated in mammalian adaptation of subtype H5N1 viruses (7–9).
Phylogenetic analysis based on the HA sequence and on complete genome sequences of HPAI (H5N1) strains grouped the 3 new isolates from Nigeria with other isolates from Europe, the Middle East, and Lagos state of Nigeria. According to recent classification by the H5N1 Evolution Working Group, the viruses belong to clade 2.2.2 (previously referred to as the European-Middle Eastern-African clade 1) (10). The Technical Appendix, shows the phylogenetic trees generated on the basis of the sequences of HA, NA, nucleocapsid protein (NP), and NS segments. On the basis of phylogenetic analyses of all 8 segments, no evidence of reassortment was observed among the newly sequenced HPAI isolates from Nigeria. Although prior reports (2,3,11) have suggested 3 independent introductions of HPAI viruses into Nigeria, our analysis of viruses in this study and in GenBank identified only 2 clades (clades 2.2.2 and 2.2.3) among the Nigeria isolates, suggesting 2 unique introductions into Nigeria.
The virulence of the HPAI virus isolated in poultry was assessed by intravenous injection of a 1:10 dilution of allantoic fluid into groups of 4-week-old SPF White Leghorn chickens, 8 per group, as previously described (12). According to results of this assay, all 3 isolates were highly pathogenic (Table); mean times to death were 1.0, 1.3, and 1.4 days, similar to times previously reported for other European-Asian lineage HPAI (H5N1) viruses (13).
To determine infectivity and pathogenicity, using a simulated natural respiratory route of exposure, we intranasally inoculated ten 4-week-old White Leghorn chickens with strain A/chicken/Nigeria/228-5/2005 (106 mean embryo infectious doses). The caged birds were inspected daily for clinical signs and death. Two chickens that were not inoculated and were maintained as negative controls were euthanized (100 mg sodium pentobarbital/kg body weight) at 0 days postinoculation (dpi). At 1 dpi, 2 of the injected birds were euthanized. On 2 dpi, postmortem examinations were performed on 2 birds that had died, and samples were collected for virus isolation and for histopathologic and immunohistochemical examination of a variety of tissues. For immunohistochemical examination, a monoclonal antibody against influenza A nucleoprotein was used as previously described (14). Virus isolation and titration were conducted with brain, heart, lung, kidney, breast muscle, oropharyngeal, and cloacal samples; embryonating chicken eggs were used (12).
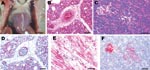
After intranasal inoculation with A/chicken/Nigeria/228-5/2005, all birds died or were euthanized for severe illness within 3 dpi (mean time to death 2.125 days). At 1 dpi, both intranasally inoculated chickens sampled were infected, as evidenced by low virus titers in oropharyngeal swabs; however, only 1 chicken had systemic infection with moderate titers of virus in tissues (Table). Histologically, both chickens lacked lesions, but the spleen of 1 chicken had a few avian influenza virus–positive histiocytes. The intranasally inoculated chickens that died and were necropsied on 2 dpi had exhibited mild to moderate listlessness, a hunched posture, and partial closure of eyelids a few hours before death. Gross lesions included scattered petechia in epicardial fat within the coronary groove (Figure, panel A), a few petechia in mucosa of the ventriculi (gizzards), slight increase in pericardial fluid, and swollen kidneys. Swabs from respiratory and alimentary tracts and the tissues had high titers of the virus (Table). Histologically, tissues with lesions coincided with sites of virus replication and indicated severe systemic infection. The most severe lesions were severe interstitial pneumonia with edema (Figure, panel B), moderate to severe myocyte degeneration and necrosis in the heart, moderate nonsuppurative encephalitis, moderate necrotizing rhinitis, moderate lymphohistiocytic depletion and apoptosis in the spleen, and mild to moderate degeneration and necrosis of pancreatic acinar epithelium (Figure, panel C). Scattered lesions of necrosis and inflammation were seen in liver, cloacal bursa, thymus, proventriculus, ventriculus, and pancreatic islets. Influenza A virus was localized to necrotic cells, which most frequently included blood vessel endothelium throughout the body (Figure, panel D), cardiac myocytes (Figure, panel E), pulmonary histiocytes and heterophils (Figure, panel D), neurons and glial cells in the brain, splenic histiocytes and cellular debris, and renal tubular epithelium. Virus was less frequently identified in Kupffer cells and hepatocytes; histiocytes in lamina propria of alimentary tract, thymus, and cloacal bursa; pancreatic acinar and islet epithelium (Figure, panel F); proventricular epithelium; and bone marrow myeloid cells.
The 3 early HPAI (H5N1) isolates from Nigeria, which belonged to clade 2.2.2, produced in chickens a systemic infection characterized by virus replication and associated necrotic and inflammatory lesions in critical internal organs such as the heart, brain, and lungs. A prominent vascular tropism of the virus was evidenced by widespread replication in blood vessel endothelium throughout the body and is typical of other HPAI viruses (H5N1) of the Asian lineage.
Dr Aiki-Raji performs virus surveillance and pathogenesis research at the poultry clinic of the University of Ibadan Veterinary Teaching Hospital.
Acknowledgments
We thank Joan Beck, Kira Moresco, and James Doster for technical assistance.
This work was supported by National Institutes of Health grants P01 AI058113 and U54 AI057158 (Northeast Biodefense Center-Lipkin), and US Department of Agriculture, Agricultural Research Service, Current Research Information System project 6612-32000-048-00D. P.V.A. was supported by a fellowship awarded by AI057158 (Northeast Biodefense Center-Lipkin).
References
- The World Health Organization Global Influenza Program Surveillance Network. Evolution of H5N1 avian influenza viruses in Asia. Emerg Infect Dis. 2005;11:1515–21.PubMedGoogle Scholar
- Ducatez MF, Olinger CM, Owoade AA, De Landtsheer S, Ammerlaan W, Niesters HG, Avian flu: multiple introductions of H5N1 in Nigeria. Nature. 2006;442:37. DOIPubMedGoogle Scholar
- Ducatez MF, Olinger CM, Owoade AA, Tarnagda Z, Tahita MC, Sow A, Molecular and antigenic evolution and geographical spread of H5N1 highly pathogenic avian influenza viruses in western Africa. J Gen Virol. 2007;88:2297–306. DOIPubMedGoogle Scholar
- Ives JA, Carr JA, Mendel DB, Tai CY, Lambkin R, Kelly L, The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res. 2002;55:307–17. DOIPubMedGoogle Scholar
- Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A. 2008;105:4381–6. DOIPubMedGoogle Scholar
- Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–80. DOIPubMedGoogle Scholar
- Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–2. DOIPubMedGoogle Scholar
- Shinya K, Hamm S, Hatta M, Ito H, Ito T, Kawaoka Y. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology. 2004;320:258–66. DOIPubMedGoogle Scholar
- Chen H, Li Y, Li Z, Shi J, Shinya K, Deng G, Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J Virol. 2006;80:5976–83. DOIPubMedGoogle Scholar
- Salzberg SL. Genome analysis linking recent European and African influenza (H5N1) viruses. Emerg Infect Dis. 2007;13:713–8.PubMedGoogle Scholar
- Ducatez MF, Tarnagda Z, Tahita MC, Sow A, de Landtsheer S, Londt BZ, Genetic characterization of HPAI (H5N1) viruses from poultry and wild vultures, Burkina Faso. Emerg Infect Dis. 2007;13:611–3.PubMedGoogle Scholar
- Swayne DE, Senne DA, Beard CW. Isolation and identification of avian pathogens. In: Swayne DE, Jackwood MW, Pearson JE, Reed WM, editors. Influenza. 4th ed. Kennett Square (PA): American Association of Avian Pathologists; 1998. p. 150–5.
- Swayne DE, Pantin-Jackwood M. Pathogenicity of avian influenza viruses in poultry. Dev Biol (Basel). 2006;124:61–7.PubMedGoogle Scholar
- Swayne DE. Pathobiology of H5N2 Mexican avian influenza virus infections of chickens. Vet Pathol. 1997;34:557–67.PubMedGoogle Scholar
Figure
Table
Cite This ArticleTable of Contents – Volume 14, Number 11—November 2008
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Christopher F. Basler, Department of Microbiology, Box 1124, Mount Sinai School of Medicine, 1 Gustave L. Levy Place, New York, NY 10029, USA;
Top