Volume 15, Number 2—February 2009
Research
Simian T-Lymphotropic Virus Diversity among Nonhuman Primates, Cameroon
Figure 6
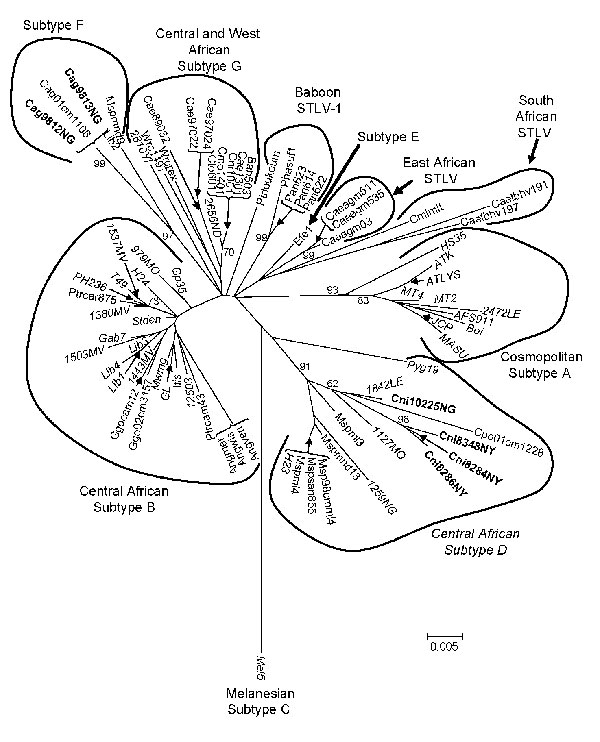
Figure 6. Inferred phylogenetic relationships of primate T-lymphotropic virus (PTLV)–1 long terminal repeat (LTR) sequences by neighbor-joining analysis. Sequences from wild nonhuman primates (NHPs) in Cameroon generated in the current study are in boldface. Human T-lymphotropic virus–1 seqences are italicized. Support for the branching order was determined by 1,000 bootstrap replicates; only values >60% are shown. STLV, simian T-lymphotropic virus. See Figure 2 legend for additional abbreviations.
1Current affiliation: Stanford University, Stanford, California, USA.
2Current affiliation: University of Pittsburgh Graduate School of Public Health, Pittsburgh, Pennsylvania, USA.
Page created: December 08, 2010
Page updated: December 08, 2010
Page reviewed: December 08, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.