Volume 15, Number 6—June 2009
Research
Lineage 2 West Nile Virus as Cause of Fatal Neurologic Disease in Horses, South Africa
Figure 2
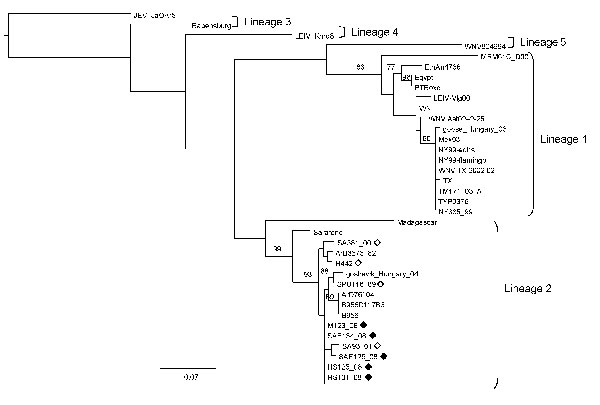
Figure 2. Maximum-likelihood comparison of the partial NS5 gene of West Nile virus (WNV) strains identified in horses in South Africa in 2008 with representative sequences of other WNV lineages. Bootstrap statistics are shown on the branches; only values >70% are included. Scale bar indicates 0.07 nt changes. Japanese encephalitis virus (JEV) was used as an outgroup. Black diamonds, WNV strains identified in horses in South Africa in the present study; white diamonds, WNV strains isolated from humans in South Africa in previous years. WNV strains and accession numbers and origin: HS123_08, FJ464376, South Africa; HS125_08, FJ464377, South Africa; HS101_08, FJ464378, South Africa; SAE126_08, FJ464379, South Africa; SAE134_08, FJ464380, South Africa; SA381_00, EF429199, South Africa; SA93_01, EF429198, South Africa; SPU116_89, EF429197, South Africa; goshawk_Hungary_04, DQ116961, Hungary; B956 polyprotein gene-1937, AY532665, Uganda; B956 117B3, M12294, Uganda; ArD76104, DQ318019, Senegal; H442, EF429200, South Africa; ArB3573_82, DQ318020, Central African Republic; Sarafend, AY688948, Uganda; Madagascar AnMg798, DQ176636, Madagascar; PTRoxo, AM404308, Portugal; Egypt101, AF260968, Egypt; EthAn4766, AY603654, Ethiopia; Kunjin MRM61C, D00256, Australia; WNV Italy 1998 equine, AF404757, Italy; WNV Ast02–2-25, DQ374653, Russia; LEIV-Vlg00–27924, AY278442, Russia; LEIV-Krnd88–190, AY277251, Russia; goose_Hungary_03, DQ118127, Hungary; NY385_99, DQ211652 NY, USA; NY99-eqhs, AF260967 NY, USA; NY99-flamingo382–99, AF196835 NY, USA; TYP9376 NY385_99, AY848697 NY, USA; WNV TX 2002 02, DQ164206 TX, USA; TM171_03, AY371271, Mexico; Mex03, AY660002, Mexico; TX 2002-HC, DQ176637 TX, USA; WNV804994, DQ256376, India; Rabensburg 97–103, AY765264, Czech Republic; JEV JaOAr5982, M18370, Japan.