Volume 16, Number 6—June 2010
Dispatch
Xenotropic Murine Leukemia Virus–related Gammaretrovirus in Respiratory Tract
Cite This Article
Citation for Media
Abstract
Xenotropic murine leukemia virus–related gammaretrovirus (XMRV) has been recently associated with prostate cancer and chronic fatigue syndrome. To identify nucleic acid sequences, we examined respiratory secretions by using PCR. XMRV-specific sequences were detected in 2%–3% of samples from 168 immunocompetent carriers and ≈10% of samples from 161 immunocompromised patients.
Xenotropic murine leukemia virus–related gammaretrovirus (XMRV) was originally discovered in tissue from patients with familial prostate cancer homozygous for a missense mutation in the RNase L gene, R462Q (1). Detection of viral nucleic acid in tissue sections of cancerous prostate glands and cloning of the viral integration sites confirmed XMRV as a bona fide human infection with a murine leukemia virus–related retrovirus (1). Whether XMRV is actively involved in prostate cancer tumorigenesis or whether it is just a bystander virus (2,3) remains unclear.
On the basis of its close homology (up to 94% nt identity) to endogenous and exogenous full-length sequences from Mus musculus mice (1), XMRV most likely originated in mice, although they are probably not the current reservoir of infection (4). Recent findings of XMRV sequences in up to 67% of peripheral blood mononuclear cells (PBMCs) of patients with chronic fatigue syndrome and in 3.4% of PBMCs of healthy controls raise the question whether XMRV could be a blood-borne pathogen (5). However, the finding of XMRV in PBMCs from patients with chronic fatigue syndrome is controversial because multiple studies in Europe have failed to detect XMRV (6–8). Similarly, frequency of XMRV in prostate cancer samples ranges from 0 to 23%, depending on geographic restriction of the virus or, more likely, diagnostic techniques used (PCR, quantitative PCR, immunohistochemistry) (1–3,9,10). Indirect evidence has suggested sexual transmission (9). Questions remain about worldwide distribution, host range, transmission routes, and organ tropism of the virus. To begin to answer some of them, we looked for XMRV in respiratory samples from 267 patients with respiratory tract infection (RTI) and 62 healthy persons.
During 2006–2009, the 267 samples were collected from 3 groups of patients (Table). Group 1 comprised patients who had traveled from Asia to Germany; location of their permanent residency was unknown. Groups 2 and 3 and the control group comprised only persons from northern Germany. From group 1, a total of 75 sputum and nasal swab specimens were collected from patients who had unconnected cases of RTI and who had recently traveled by air (11). From group 2, a total of 31 bronchoalveolar lavage (BAL) samples were collected from patients with chronic obstructive pulmonary disease (defined by a forced expiratory volume in 1 second/forced vital capacity <70% and forced expiratory volume in 1 second <80% of the predicted value) who had signs of RTI. From group 3, a total of 161 BAL and tracheal secretion samples were collected from patients with severe RTI and immunosuppression as a result of solid organ or bone marrow transplantation. From the control group, throat swabs were collected from 52 healthy persons and BAL samples were collected from 10 healthy volunteers who had no signs of RTI and no known underlying disease.
All samples were analyzed by culture for pathogenic bacteria and fungi and by PCR for rhinoviruses, adenoviruses, enteroviruses, influenza viruses A and B, parainfluenza viruses 1–3, respiratory syncytial virus, cytomegalovirus, Epstein-Barr virus, and human metapneumovirus. All samples were tested in duplicates obtained by individual RNA extractions. XMRV RNA was reverse transcribed from total RNA, after which nested PCR or real-time PCR were conducted as recently described (1,12). No serum samples were available from these patients to confirm the results by serologic testing.
For group 1, XMRV-specific sequences were detected with relatively low frequency (2.3%). For group 2, XMRV-specific sequences were amplified in 1 BAL sample, which was also positive for Staphylococcus aureus by routine culture methods. For group 3, XMRV-specific sequences were detected at a frequency of 9.9%, which was significantly higher than that for the healthy control group (3.2%) at the 90% confidence level but not at the 95% level (p = 0.078, 1 sample t-test). Of 16 group 3 samples, 10 showed no signs of co-infection. The remaining 6 samples showed co-infection with rhinovirus or adenovirus (1 sample each); S. aureus (3 samples); or mixed infection with pathogenic fungi, Candida albicans and Asperigillus fumigatus (1 sample).
All samples that were positive for XMRV by gag-nested PCR, together with a set of those that were negative for XMRV, were retested by real-time PCR. Results showed low XMRV RNA concentrations, 103 –104/mL of specimen.
To confirm the validity of XMRV detection, a subset of 6 specimens (3 XMRV positive and 3 XMRV negative) were tested by using an alternative PCR assay for viral RNA (3) and a C-Type RT Activity Kit (Cavidi, Uppsala, Sweden) for type C reverse-transcription activity. XMRV sequences from alternative targets in the gag and env regions were confirmed in 2 of the 3 XMRV-positive samples but in none of the controls. One XMRV-positive BAL specimen showed an 8-fold increase above background of specific type C retroviral reverse-transcriptase activity, suggesting presence of active type C retrovirus within this sample. This assay is substantially less sensitive than reverse transcription–PCR.
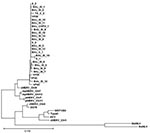
All XMRV gag sequences (390-bp fragment) were 98%–99% identical to previously published XMRV sequences from persons with prostate cancer (1,2). Phylogenetic analysis showed close clustering (Figure).
XMRV, originally identified in RNase L–deficient patients with familial prostate cancer, has gained interest since recent work showed its protein expression in as many as 23% of prostate cancer cases (10) and XMRV-specific sequences were detected in PBMCs of 67% patients with chronic fatigue syndrome (5). These results, however, could not be confirmed by others (6–8). Both studies also detected XMRV protein or sequences in their control cohorts with frequencies of 6% and 4%, respectively.
Among the most pressing information gaps with regard to XMRV is its preferred route of transmission. Detection of XMRV in PBMCs and plasma of patients with chronic fatigue syndrome raises the possibility of blood-borne transmission; sexual transmission has also been hypothesized on the basis of indirect evidence (5,9). We detected XMRV in respiratory secretions of immunocompetent patients with and without RTI at a frequency of ≈3.2%, which is in good concordance with the recently reported prevalence in the general population of up to 4% (5). Frequency of XMRV detection in group 1 patients (2.25%) was comparable to that of human metapneumovirus and rhinovirus within this group and considerably less frequent than that of parainfluenzavirus (15.5%) or influenza A virus (7.6%) detection (11).
Our findings indicate that XMRV or virus-infected cells might be carried in and transmitted by the respiratory tract. Attempts to isolate infectious virus from XMRV sequence–positive respiratory samples failed, possibly because of inadequate storage of samples before virus culturing attempts or relatively low copy numbers of the virus within the samples. Thus, whether the respiratory tract serves as a putative transmission route for XMRV cannot be determined at this time. The observed increase in prevalence among immunosuppressed patients with RTI suggests that XMRV might be reactivated in absence of an efficient antiviral defense. Together with earlier observations on increased XMRV replication in RNase L–deficient cells (1,12), this finding implies that the immune system plays a role in controlling XMRV replication. It remains unknown whether immunosuppression predisposes a patient to secrete infectious XMRV from the respiratory tract or whether presence of virus might be meaningless for epidemiology in a way similar to HIV-1 (15). Future studies should address whether the respiratory tract might serve as a source of XMRV infection or whether immunosuppression might cause an increased risk for primary infection.
Dr Fischer works as a group leader at the Institute for Medical Microbiology and Virology at the University Medical Center Hamburg-Eppendorf. Her main research interests are emerging viruses, in particular the gammaretrovirus XMRV.
Acknowledgment
This study was supported by the Werner Otto Stiftung grant no. 4/69 to N.F. The study was approved by the ethics committee at the board of physicians of the Free and Hanseatic City of Hamburg (No.WF-005/09).
References
- Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. DOIPubMedGoogle Scholar
- Fischer N, Hellwinkel O, Schulz C, Chun FK, Huland H, Aepfelbacher M, Prevalence of human gammaretrovirus XMRV in sporadic prostate cancer. J Clin Virol. 2008;43:277–83. DOIPubMedGoogle Scholar
- Hohn O, Krause H, Barbarotto P, Niederstadt L, Beimforde N, Denner J, Lack of evidence for xenotropic murine leukemia virus–related virus (XMRV) in German prostate cancer patients. Retrovirology. 2009;6:92. DOIPubMedGoogle Scholar
- Stieler KSC, Lavanya M, Aepfelbacher M, Stocking C, Fischer N. Host range and cellular tropism of the human exogenous gammaretrovirus XMRV. Virology. 2010;399:23–30. DOIPubMedGoogle Scholar
- Lombardi VC, Ruscetti FW, Das Gupta J, Pfost MA, Hagen KS, Peterson DL, Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science. 2009;326:530–1. DOIPubMedGoogle Scholar
- Erlwein O, Kaye S, McClure MO, Weber J, Wills G, Collier D, Failure to detect the novel retrovirus XMRV in chronic fatigue syndrome. PLoS ONE. 2010;5:e8519. DOIPubMedGoogle Scholar
- Groom HC, Boucherit VC, Makinson K, Randal E, Baptista S, Hagan S, Absence of xenotropic murine leukaemia virus–related virus in UK patients with chronic fatigue syndrome. Retrovirology. 2010;7:10. DOIPubMedGoogle Scholar
- van Kuppeveld FJ, de Jong AS, Lanke KH, Verhaegh GW, Melchers WJ, Swanink CM, Prevalence of xenotropic murine leukaemia virus–related virus in patients with chronic fatigue syndrome in the Netherlands: retrospective analysis of samples from an established cohort. BMJ. 2010;340:c1018. DOIPubMedGoogle Scholar
- Hong S, Klein EA, Das Gupta J, Hanke K, Weight CJ, Nguyen C, Fibrils of prostatic acid phosphatase fragments boost infections with XMRV (xenotropic murine leukemia virus–related virus), a human retrovirus associated with prostate cancer. J Virol. 2009;83:6995–7003. DOIPubMedGoogle Scholar
- Schlaberg R, Choe D, Brown KR, Thaker HM, Singh IR. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc Natl Acad Sci U S A. 2009;106:16351–6. DOIPubMedGoogle Scholar
- Luna LK, Panning M, Grywna K, Pfefferle S, Drosten C. Spectrum of viruses and atypical bacteria in intercontinental air travelers with symptoms of acute respiratory infection. J Infect Dis. 2007;195:675–9. DOIPubMedGoogle Scholar
- Dong B, Kim S, Hong S, Das Gupta J, Malathi K, Klein EA, An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc Natl Acad Sci U S A. 2007;104:1655–60. DOIPubMedGoogle Scholar
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–5. DOIPubMedGoogle Scholar
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. DOIPubMedGoogle Scholar
- Wood KL, Chaiyarit P, Day RB, Wang Y, Schnizlein-Bick CT, Gregory RL, Measurements of HIV viral loads from different levels of the respiratory tract. Chest. 2003;124:536–42. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleTable of Contents – Volume 16, Number 6—June 2010
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Nicole Fischer, Institute for Medical Microbiology and Virology, University Medical Center Hamburg-Eppendorf, Martinistrasse 52, 20246 Hamburg, Germany
Top