Volume 16, Number 8—August 2010
Dispatch
Novel Mycobacterium tuberculosis Complex Pathogen, M. mungi
Cite This Article
Citation for Media
Abstract
Seven outbreaks involving increasing numbers of banded mongoose troops and high death rates have been documented. We identified a Mycobacterium tuberculosis complex pathogen, M. mungi sp. nov., as the causative agent among banded mongooses that live near humans in Chobe District, Botswana. Host spectrum and transmission dynamics remain unknown.
A previously unidentified Mycobacterium tuberculosis complex pathogen has emerged in banded mongooses (Mungos mungo) in Botswana; we named the pathogen mongoose bacillus, or M. mungi sp. nov. This pathogen causes high mortality rates among banded mongooses that live in close association with humans because these animals live in human-made structures and scavenge human waste, including feces.
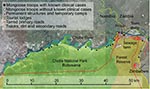
Banded mongooses are social, fossorial, viverids that feed on invertebrates and small mammals including subterranean species (1). We initially identified tuberculosis (TB) disease in banded mongooses in 2000. The outbreak appeared to spread as a point-source infection between mongoose troops living in close association with humans and human waste; infection spread through towns and the associated national park (2). During 2000–2010, a total of 7 outbreaks occurred (increasing in duration), mongoose troop involvement increased, and the spatial and temporal connection between cases decreased. Infected mongoose troops are now widely identified across the landscape, including protected areas and urban centers (Figure 1), and high mortality rates threaten the survival of smaller troops. In this study area of Chobe District, Botswana, TB has been identified in only humans and mongooses. Strain assessment of human TB has not been conducted; the full host spectrum and transmission dynamics of this pathogen, currently unknown, are the focus of our ongoing research.
During 2000–2010 in Chobe District, Botswana, we performed 38 necropsies on macroscopically TB-positive mongooses, of which 18 were further evaluated and TB was confirmed by histopathologic examination. An in-depth histologic evaluation was performed on a subsample of 8 of these animals from the 2008 outbreak. The most striking feature identified in the sick mongooses was anorexia, followed by nasal distortion and, less commonly, erosions of the nasal planum with involvement of the hard palate. For 7 of the 8 TB-positive animals examined intensively, macroscopic lesions were noted on the nasal planum. Histologic examination detected unequivocal TB lesions in the skin of the nose and the anterior nasal mucosa. Our findings suggested entry of the organism through erosions on the nasal planum, perhaps in association with abrasions, which might occur during foraging. Such lesions were present in the hairless parts of the nose tip of most TB-infected mongooses. Furthermore, granulomatous inflammation and mycobacterial organisms were found in the dermis of the skin directly below these erosions. Inflammation and organisms were present in some cases in the nasal mucosa, but erosion was not found. Thus, organisms could not have been in the lumen of the nasal cavity. This finding is consistent with pneumonic TB being present in only a few advanced cases of disseminated TB. This pattern was consistent among all animals examined postmortem during the study period. Histologically, the TB pneumonia was determined to be hematogenous rather than bronchogenous (i.e., by inhalation); thus, no evidence for aerosol transmission was found. Rather, pathogen invasion appears to have occurred through the nasal planum of the mongoose, and hematogenous or lymphatic spread through the body was a strikingly unique feature of this particular M. tuberculosis complex organism.
Samples from histologically positive mongooses were positive by PCR for the MPB70 target, IS6110 element, and 16S rDNA, indicating that the infective organism was a member of the M. tuberculosis complex (3,4). Samples were further evaluated with an M. tuberculosis complex–specific multiplex PCR (5), which provided distinct results, differing clearly from those for other members of the M. tuberculosis complex (Table 1).
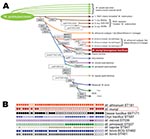
The gyrB gene (encoding for gyrase B) sequence, used to identify M. tuberculosis complex member–specific sequence single-nucleotide polymorphisms (SNPs) (6), identified the position of the organism as being situated between dassie bacillus and M. africanum subtype 1(a) and showed no detectable new SNPs (Figure 2, panel A). Amplification of RD701 and RD702 and lack of SNPs in rpoB and hsp65 genes demonstrated that this organism was not a member of the M. africanum subtype 1(a) sublineage (6,11) (Figure 2, panel A).
Three markers were evaluated to definitively exclude the organism from being dassie bacillus: N-RD25das deletion, RD1das deletion, and SNP 389 in the gene Rv0911 (6). The N-RD25das amplification gave the right product for the presence of a deletion in this region, and further sequencing confirmed that it contained a deletion in the same position as N-RD25das; however, sequencing of Rv0911 showed no SNP at position 389, indicating that this organism was not dassie bacillus. As a final test, we amplified the RD1das region but were unable to amplify a product from the mongoose isolates. We redesigned primers to amplify a smaller region of different diagnostic sizes (248 bp when RD1das is deleted and 318 bp when RD1 is intact) but still had no amplification. This finding indicates that the RD1 region is deleted in this organism but that the deletion is larger than that of the dassie bacillus.
We then used spoligotyping analysis (12) to further evaluate mongoose samples and identified a unique spoligotype pattern with no known matches in the international spoligotyping database SpolDB4 (9) or the M. bovis–specific spoligotype database (www.Mbovis.org) (Figure 2, panel B). This pattern was constant during 2000–2009 in different mongoose troops and locations (Technical Appendix). This unique spoligotyping pattern will enable identification of M. mungi in future TB surveillance programs.
For these same isolates, the full set of 24 mycobacterial interspersed repetitive unit–variable number tandem repeats (13) identified a pattern that was unique compared with others in the international database at www.miru-vntrplus.org (Table 2). Our examination also included dassie bacillus, which had not been previously analyzed. Evidence of multiple M. mungi substrains circulating between years and within social groups (6601B and 6600B) in the same outbreak year (Table 2) suggests complexity in M. mungi transmission and potential evolution of the organism over the past decade.
This newly identified mycobacterial pathogen has many unique ecologic characteristics that set it apart from other members of the M. tuberculosis complex. First, it causes high numbers of deaths of banded mongooses, threatening local extinction of smaller social groups. Second, rather than having a primary respiratory transmission route with direct transmission between individuals, as is characteristic of other M. tuberculosis complex species, M. mungi appears to infect banded mongooses by means of a nonrespiratory route through the nasal planum, suggestive of environmental transmission. Third, the time from clinical presentation to death for affected mongooses is generally short (2–3 months) compared with that for other M. tuberculosis complex pathogens (more chronic infection, can take years to progress to death). Acute illness and high mortality rates, as seen in banded mongooses with M. mungi infection, have been associated with extremely isolated human communities newly exposed to TB (14).
Conventional laboratory culture, biochemical testing, and a limited molecular evaluation were insufficient for differentiating M. mungi from M. tuberculosis (2). Organism differentiation required an extensive suite of additional molecular assessments not available at that time, thus underscoring the difficulty of diagnosing M. tuberculosis complex agents correctly and the inability of most national health laboratories to do so. The fact that new host-adapted M. tuberculosis complex species continue to be identified illustrates the diversity within the M. tuberculosis complex and stresses the need for sensitive techniques for species differentiation. The identification of this previously unknown pathogen within the M. tuberculosis complex identifies new concerns for human and animal health and illustrates the continuing scope of the threat posed by TB pathogens.
Dr Alexander is a researcher at Virginia Polytechnic Institute and State University. Her research is directed at understanding the ecology of infectious disease at the human–animal interface in Africa.
Acknowledgments
We acknowledge the assistance of the Botswana Department of Wildlife and National Parks. We also thank Tara Craig and Tiny Hlokwe for their assistance with this project.
The work was funded through a grant from National Geographic Society, WildiZe Foundation, and Virginia Polytechnic Institute and State University.
References
- Alexander K, Pleydell E, Williams M, Lane E, Nyange J, Michel A. Mycobacterium tuberculosis: an emerging disease of free-ranging wildlife. Emerg Infect Dis. 2002;8:598–601.PubMedGoogle Scholar
- Liébana E, Aranaz A, Francis B, Cousins D. Assessment of genetic markers for species differentiation within the Mycobacterium tuberculosis complex. J Clin Microbiol. 1996;34:933.PubMedGoogle Scholar
- Harmsen D, Dostal S, Roth A, Niemann S, Rothgänger J, Sammeth M, RIDOM: comprehensive and public sequence database for identification of Mycobacterium species. BMC Infect Dis. 2003;3:26. DOIPubMedGoogle Scholar
- Warren R, Gey van Pittius N, Barnard M, Hesseling A, Engelke E, De Kock M, Differentiation of Mycobacterium tuberculosis complex by PCR amplification of genomic regions of difference. Int J Tuberc Lung Dis. 2006;10:818–22.PubMedGoogle Scholar
- Huard R, Fabre M, de Haas P, Claudio Oliveira Lazzarini L, van Soolingen D, Cousins D, Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J Bacteriol. 2006;188:4271. DOIPubMedGoogle Scholar
- Gordon S, Bottai D, Simeone R, Stinear T, Brosch R. Pathogenicity in the tubercle bacillus: molecular and evolutionary determinants. Bioessays. 2009; 31.PubMedGoogle Scholar
- Kremer K, Van Soolingen D, Frothingham R, Haas W, Hermans P, Martin C, Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607.PubMedGoogle Scholar
- Brudey K, Driscoll J, Rigouts L, Prodinger W, Gori A, Al-Hajoj S, Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB 4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:23. DOIPubMedGoogle Scholar
- van Soolingen D, van der Zanden A, de Haas P, Noordhoek G, Kiers A, Foudraine N, Diagnosis of Mycobacterium microti infections among humans by using novel genetic markers. J Clin Microbiol. 1998;36:1840.PubMedGoogle Scholar
- Mostowy S, Onipede A, Gagneux S, Niemann S, Kremer K, Desmond E, Genomic analysis distinguishes Mycobacterium africanum. J Clin Microbiol. 2004;42:3594. DOIPubMedGoogle Scholar
- Kamerbeek J, Schouls L, Kolk A, Van Agterveld M, Van Soolingen D, Kuijper S, Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907.PubMedGoogle Scholar
- Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, Proposal for standardization of optimized mycobacterial interspersed repetitive unit–variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498. DOIPubMedGoogle Scholar
- Sousa A, Salem J, Lee F, Vercosa M, Cruau P, Bloom B, An epidemic of tuberculosis with a high rate of tuberculin anergy among a population previously unexposed to tuberculosis, the Yanomami Indians of the Brazilian Amazon. Proc Natl Acad Sci U S A. 1997;94:13227–23. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 16, Number 8—August 2010
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Kathleen A. Alexander, Virginia Polytechnic Institute and State University, Fisheries and Wildlife Sciences, 152 Cheatham Hall (0321), Blacksburg, VA 24061, USA
Top