Volume 21, Number 6—June 2015
Dispatch
KPC and NDM-1 Genes in Related Enterobacteriaceae Strains and Plasmids from Pakistan and the United States
Cite This Article
Citation for Media
Abstract
To characterize the genomic context of New Delhi metallo-β-lactamase-1 (NDM-1) and Klebsiella pneumoniae carbapenemase (KPC), we sequenced 78 Enterobacteriaceae isolates from Pakistan and the United States encoding KPC, NDM-1, or no carbapenemase. High similarities of the results indicate rapid spread of carbapenem resistance between strains, including globally disseminated pathogens.
Pathogenic Enterobacteriaceae, including Escherichia coli and Klebsiella pneumoniae, are major causes of multidrug-resistant (MDR) infections in hospitals worldwide. These pathogens have recently been shown to have acquired resistance to carbapenems, and the US Centers for Disease Control and Prevention identified carbapenem-resistant Enterobacteriaceae as of the 3 most urgent MDR threats (1). Among the Enterobacteriaceae, β-lactam resistance, including carbapenem resistance, is primarily caused by enzymatic degradation by β-lactamases. Two carbapenemase subclasses are especially problematic: Klebsiella pneumoniae carbapenemase (KPC) and New Delhi metallo-β-lactamase-1 (NDM-1). KPC, identified in 2001 (2), has become endemic to several noncontiguous areas of the world, including the United States, Israel, Greece, South America, and China (3). NDM-1 was first described in 2008, although retrospective studies identified NDM-1 from 2006 (4) and is abundant in New Delhi water samples (5). Most patients from whom NDM-1 is isolated have an epidemiologic link to the Indian subcontinent, but NDM-1 has also recently become endemic to the Balkans and Middle East (6).
The spread of antibiotic resistance genes such as NDM-1 and KPC is facilitated by horizontal gene transfer (HGT) between bacteria (7). Among globally disseminated pathogens, HGT facilitates combination of the most effective antibiotic resistance genes from diverse geographies into multidrug resistance plasmids that spread between strains. Recombination and transposition have created populations of these plasmids that have related architectures but vary in their composition of antibiotic drug resistance cassettes (8). This effect has enabled both KPC and NDM-1 to rapidly expand within the Enterobacteriaceae and other proteobacterial pathogens, such as Acinetobacter baumanii (9,10). Antibiotic resistance genes can also spread through clonal expansion in successful pathogenic strains, for example, KPC in K. pneumoniae sequence type (ST) 258 (11), and the extended-spectrum β-lactamase CTX-M-15 in E. coli ST131 (12). Both HGT and clonal expansion have enabled KPC and NDM-1 to rapidly spread to distant locations after their emergence (6,8).
The similarities in the spread and resistance spectra of KPC and NDM-1 (both provide resistance to nearly all β-lactam antimicrobial drugs) leads to the hypothesis that similar mobile elements will make both genes available to similar pathogen populations. We tested this hypothesis by examining clinical Enterobacteriaceae isolates from Pakistan and the United States encoding NDM-1, KPC, or no carbapenemase.
We collected 450 bacterial isolates (including 195 Enterobacteriaceae) in Pakistan during February 2012–March 2013 from Pakistan Railway General Hospital in Rawalpindi and the Pakistan Institute of Medical Sciences in Islamabad. From this collection, we randomly selected 55 Enterobacteriaceae isolates for whole-genome sequencing. We then selected 23 isolates from samples collected in the United States during January 2010–June 2013 from patients in Barnes Jewish Hospital in St. Louis, Missouri, that had similar proportions of β-lactam susceptibility and resistance to the isolates collected in Pakistan for sequencing. All isolates were de-identified and retrieved from existing strain banks. The combined set included 33 E. coli, 30 K. pneumoniae, 9 Enterobacter cloacae, and 6 Enterobacter aerogenes (Technical Appendix Table 1). We extracted plasmid DNA from 9 isolates encoding NDM-1, 11 isolates encoding KPC, and 3 isolates encoding CTX-M-15 and performed shotgun sequencing on those plasmid preparations. Detailed methods are described in the Technical Appendix.
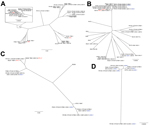
Using antibiotic resistance gene predictions from the Resfams database (13) and core genome alignment, we constructed a phylogenetic tree for each species in our set, overlaid by the β-lactamases encoded by each isolate (Figure 1). Isolates from both locations were found to be members of the same subspecies clades (Technical Appendix Figure 1) and to contain similar repertoires of β-lactamases (Figure 1), indicating that geography is not a discriminating variable for these isolates. Many of these isolates were also MDR: resistance to ciprofloxacin, trimethoprim/sulfamethoxazole, gentamicin, doxycycline, and chloramphenicol occurred in 63%, 65%, 45%, 54%, and 56% of isolates, respectively. As expected from results of previous work (8), E. coli ST131 isolates had high rates of CTX-M carriage (82%; Figure 1, panel A) and ciprofloxacin resistance (100%).
The variety of strains that we discovered encoding KPC and NDM-1 is consistent with existing evidence that HGT is a major factor in their spread. All KPC genes were proximal to Tn4401 and all NDM-1 genes were carried on ISAba125, mobile elements with which each gene has respectively been previously associated (14). We observed multiple examples of NDM-1 within the K. pneumoniae ST11 clade (15) (Figure 1, panel B; Technical Appendix Figure 1, panel B), a close relative of ST258. This association could be caused by clonal expansion or multiple HGT events and emphasizes that lineages known to encode KPC are now also acquiring NDM-1. We also observed high rates of NDM-1 carriage in Enterobacter isolates (Figure 1, panels C and D), which in general showed a high number (maximum 8) and wide variety of β-lactamases. These isolates were also MDR: 57% of the Enterobacter isolates were resistant to all or all but 1 of the antimicrobial drugs tested. At best, these Enterobacter strains are a reservoir for resistance in Pakistan; at worst, they are the vanguard of an expansion of carbapenem-resistant Enterobacter infections.
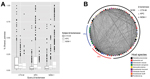
Previous observations have predominantly found KPC and NDM-1 to be expressed from plasmids (6,11). To characterize the sequence similarity of plasmids within the NDM- and KPC-carrying plasmid populations, we purified and sequenced plasmid DNA from 9 isolates encoding NDM-1, 11 encoding KPC, and 3 encoding CTX-M-15. Sequencing showed that these plasmids include representatives from IncHI2, IncY, IncN, IncFIA, IncFIB, IncFIC, and IncI1 incompatibility groups. Using reciprocal BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) alignment between each pair of plasmid preparations, we calculated the percentage of each plasmid shared using a 99% identity threshold. We performed this same analysis for all sequenced plasmids containing NDM-1, KPC, or CTX-M available in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov) together with our set (Figure 2) and separately (Technical Appendix Figure 2). Certain components, primarily mobile elements, were abundant within these plasmids: the average plasmid shared 500 contiguous bases with 58 of the other plasmids; however, median BLAST identity for this pairwise comparison was <12%, even when considering plasmids with the same β-lactamase, suggesting that both carbapenemases exist within a variety of plasmid configurations.
To visualize this comparison of carbapenemase plasmids, we generated a network diagram in which each node represented a plasmid and each line represented shared sequence between 2 plasmids (Figure 2, panel B). Node size and line width correlate to the number of nucleotides contained in the plasmid or sharing interaction. This visualization shows the abundant small, shared regions that exist between most plasmid pairs, represented as thin background lines. This visualization also highlights the larger shared regions that indicate highly similar plasmids, represented by the few wide lines. These outliers were often between pairs of plasmids encoding the same β-lactamase but were also observed between NDM-1 and KPC containing plasmids (maximum 79% of smaller plasmid length).
Together, this evidence supports our hypothesis that strains and plasmids known to carry either carbapenemase also have access to the other. Given the similarity of carbapenemase-negative strains to those carrying KPC or NDM-1 and the high diversity of plasmids in which they can be found, we anticipate that global carbapenem usage will encourage HGT of both of these carbapenemases into additional strain and plasmid backgrounds. Because KPC and NDM-1 are poised to cross genetic and geographic boundaries, we recommend that hospitals routinely screen Enterobacteriaceae strains for both genes, even in regions where they are not yet endemic. We further advocate reduced carbapenem use to limit the selection for resistance against this vital antibiotic class.
Dr. Pesesky received his PhD from Washington University in St. Louis, Missouri, USA, in 2015 and is currently a postdoctoral fellow at the University of Washington, Seattle, Washington, USA. His research focuses on molecular and genomic investigations of high interest functions in bacteria.
Acknowledgments
We thank members of the Dantas lab for thoughtful discussions of the research described herein.
Research reported in this publication was supported in part by the National Institutes of Health Director’s New Innovator Award (http://commonfund.nih.gov/newinnovator/), the National Institute of Diabetes and Digestive and Kidney Diseases (http://www.niddk.nih.gov/), and the National Institute of General Medical Sciences (NIGMS: http://www.nigms.nih.gov/), of the National Institutes of Health under award numbers DP2DK098089 and R01GM099538 to G.D. M.W.P. is supported by the NIGMS Cell and Molecular Biology Training Grant (GM:007067). T.H. is supported by the Higher Education Commission of Pakistan’s International Research Support Initiative Program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. All genome and plasmid sequences were deposited into the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov) (BioProject ID PRJNA261540).
References
- Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2013 [cited 2014 Sept 15]. http://www.cdc.gov/drugresistance/threat-report-2013/index.html
- Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–61. DOIPubMedGoogle Scholar
- Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–96. DOIPubMedGoogle Scholar
- Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob Agents Chemother. 2011;55:1274–8. DOIPubMedGoogle Scholar
- Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011;11:355–62. DOIPubMedGoogle Scholar
- Dortet L, Poirel L, Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in gram-negative bacteria. Biomed Res Int. 2014;2014:249856. DOIGoogle Scholar
- Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–21. DOIPubMedGoogle Scholar
- Hawkey PM, Jones AM. The changing epidemiology of resistance. J Antimicrob Chemother. 2009;64(Suppl 1):i3–10. DOIPubMedGoogle Scholar
- Robledo IE, Aquino EE, Vazquez GJ. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob Agents Chemother. 2011;55:2968–70. DOIPubMedGoogle Scholar
- Sartor AL, Raza MW, Abbasi SA, Day KM, Perry JD, Paterson DL, Molecular epidemiology of NDM-1 producing Enterobacteriaceae and Acinetobacter baumannii isolates from Pakistan. Antimicrob Agents Chemother. 2014;58:5589–93. DOIPubMedGoogle Scholar
- Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, Carmeli Y, Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerg Infect Dis. 2010;16:1349–56. DOIPubMedGoogle Scholar
- Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A. 2014;111:5694–9. DOIPubMedGoogle Scholar
- Gibson MKFK, Dantas G. Improved annotations of antibiotic resistance functions reveals microbial resistomes cluster by ecology. ISME. 2014;106: .DOIGoogle Scholar
- Patel G, Bonomo RA. Status report on carbapenemases: challenges and prospects. Expert Rev Anti Infect Ther. 2011;9:555–70. DOIPubMedGoogle Scholar
- Pena I, Picazo JJ, Rodriguez-Avial C, Rodriguez-Avial I. Carbapenemase-producing Enterobacteriaceae in a tertiary hospital in Madrid, Spain: high percentage of colistin resistance among VIM-1-producing Klebsiella pneumoniae ST11 isolates. Int J Antimicrob Agents. 2014;43:460–4. DOIPubMedGoogle Scholar
Figures
Cite This Article1These first authors contributed equally to this article.
Table of Contents – Volume 21, Number 6—June 2015
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Gautam Dantas, Washington University in St. Louis, 4444 Forest Park Blvd, Campus Box 8510, St. Louis, MO 63108, USA
Top