Volume 23, Number 2—February 2017
Research
Highly Pathogenic Influenza A(H5Nx) Viruses with Altered H5 Receptor-Binding Specificity
Figure 3
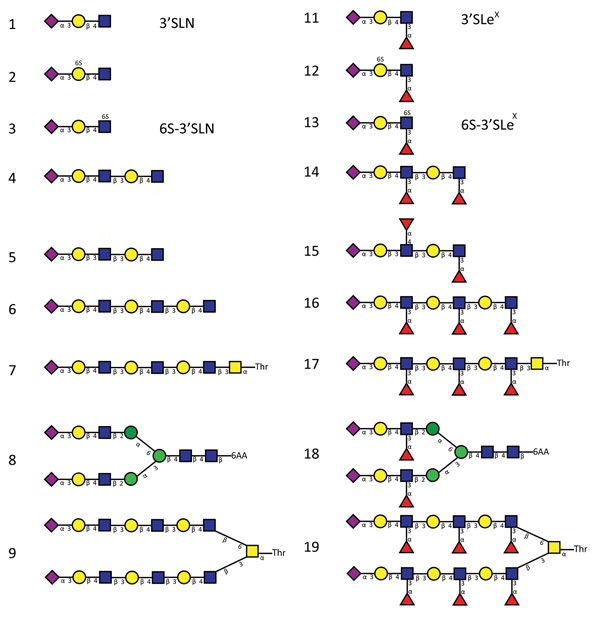
Figure 3. Glycan structures of influenza A viruses. Structures are shown for sialylated glycans present in the array in nonfucosylated (glycans 1–9) and fucosylated (glycans 11–19) forms and binding by hemagglutinins is shown in Figures 2 and 7. Glycans 1 and 11 correspond to 3′SLN (nonfucosylated glycan) and 3′SLeX (fucosylated form of 3′SLN), respectively. Similarly, glycans 3 and 13 correspond to 6-O-sulfo 3′SLN (6S-3′SLN) and 6-O-sulfo 3′SLeX (6S-3′SLeX), respectively. Blue squares, N-acetylglucosamine; yellow circles, galactose; green circles, mannose; purple diamonds, sialic acid; red triangles, fucose. H5N12.3.4, novel H5N1 virus clade 2.3.4.
1These authors contributed equally to this article.
Page created: January 17, 2017
Page updated: January 17, 2017
Page reviewed: January 17, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.