Volume 23, Number 3—March 2017
Research
Whole-Genome Analysis of Bartonella ancashensis, a Novel Pathogen Causing Verruga Peruana, Rural Ancash Region, Peru
Figure 5
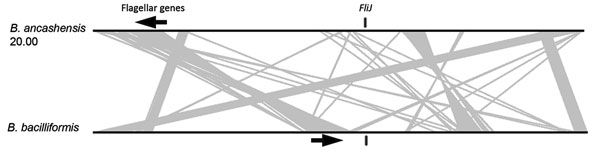
Figure 5. Genetic arrangement of the genome of B. ancashensis isolate 20.00 from a patient with verruga peruana, rural Ancash region, Peru, compared with that of B. bacilliformis KC583. Black lines indicate chromosomes and gray lines link syntenic genomic regions that are rearranged between the 2 genomes. FliJ genes are indicated by black vertical bars, and flagellar gene clusters are indicated by arrows, which indicate direction of transcription.
1Current affiliation: University of Maryland, Baltimore, Maryland, USA.
2These authors contributed equally to this article.
Page created: February 17, 2017
Page updated: February 17, 2017
Page reviewed: February 17, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.