Volume 23, Number 6—June 2017
Research
Invasive Serotype 35B Pneumococci Including an Expanding Serotype Switch Lineage, United States, 2015–2016
Figure 1
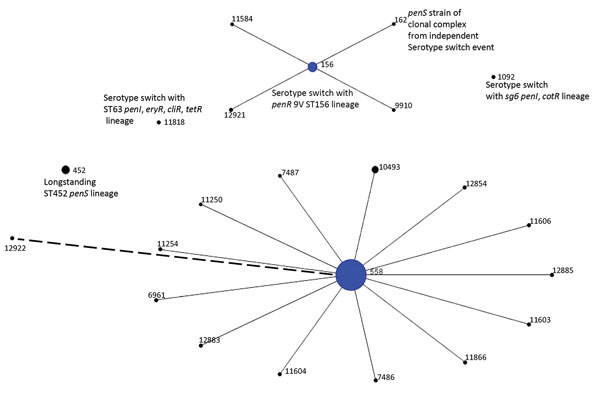
Figure 1. Population snapshot of 199 serotype 35B pneumococcal isolates obtained by ongoing Active Bacterial Core surveillance, United States, 2015–2016, configured by using eBURST (21). Diameters are proportional to number of isolates. Solid lines indicate single-locus variants, and the single dashed line indicates a double-locus variant of ST558. ST, sequence type.
References
- Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, Bennett NM, et al.; Active Bacterial Core Surveillance/Emerging Infections Program Network. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis. 2010;201:32–41. DOIPubMedGoogle Scholar
- Moore MR, Link-Gelles R, Schaffner W, Lynfield R, Lexau C, Bennett NM, et al. Effect of use of 13-valent pneumococcal conjugate vaccine in children on invasive pneumococcal disease in children and adults in the USA: analysis of multisite, population-based surveillance. Lancet Infect Dis. 2015;15:301–9. DOIPubMedGoogle Scholar
- Beall B, McEllistrem MC, Gertz RE Jr, Boxrud DJ, Besser JM, Harrison LH, et al.; Active Bacterial Core Surveillance/Emerging Infections Program Network. Emergence of a novel penicillin-nonsusceptible, invasive serotype 35B clone of Streptococcus pneumoniae within the United States. J Infect Dis. 2002;186:118–22. DOIPubMedGoogle Scholar
- Gertz RE Jr, Li Z, Pimenta FC, Jackson D, Juni BA, Lynfield R, et al. Increased penicillin-nonsusceptibility of nonvaccine serotype (other than 19A and 6A) invasive pneumococci in post 7 valent conjugate vaccine era. J Infect Dis. 2010;201:770–5. DOIPubMedGoogle Scholar
- Sharma D, Baughman W, Holst A, Thomas S, Jackson D, da Gloria Carvalho M, et al. Pneumococcal carriage and invasive disease in children before introduction of the 13-valent conjugate vaccine: comparison with the era before 7-valent conjugate vaccine. Pediatr Infect Dis J. 2013;32:e45–53. DOIPubMedGoogle Scholar
- Metcalf BJ, Gertz RE Jr, Gladstone RA, Walker H, Sherwood LK, Jackson D, et al. Active Bacterial Core surveillance team. Strain features and distributions in pneumococci from children with invasive disease before and after 13-valent conjugate vaccine implementation in the USA. Clin Microbiol Infect. 2016;22:60.e9–29. DOIGoogle Scholar
- Desai AP, Sharma D, Crispell EK, Baughman W, Thomas S, Tunali A, et al. Decline in pneumococcal nasopharyngeal carriage of vaccine serotypes after the introduction of the 13-valent pneumococcal conjugate vaccine in children in Atlanta, Georgia. Pediatr Infect Dis J. 2015;34:1168–74. DOIPubMedGoogle Scholar
- Kaur R, Casey JR, Pichichero ME. Emerging Streptococcus pneumoniae strains colonizing the nasopharynx in children after 13-valent pneumococcal conjugate vaccination in comparison to the 7-valent era, 2006–2015. Pediatr Infect Dis J. 2016;35:901–6. DOIPubMedGoogle Scholar
- Kawaguchiya M, Urushibara N, Kobayashi N. Multidrug resistance in non-PCV13 serotypes of Streptococcus pneumoniae in northern Japan, 2014. Microb Drug Resist. 2017;23:206–14. DOIPubMedGoogle Scholar
- McElligott M, Vickers I, Meehan M, Cafferkey M, Cunney R, Humphreys H. Noninvasive pneumococcal clones associated with antimicrobial nonsusceptibility isolated from children in the era of conjugate vaccines. Antimicrob Agents Chemother. 2015;59:5761–7. DOIPubMedGoogle Scholar
- Golden AR, Adam HJ, Gilmour MW, Baxter MR, Martin I, Nichol KA, et al. Assessment of multidrug resistance, clonality and virulence in non-PCV-13 Streptococcus pneumoniae serotypes in Canada, 2011-13. J Antimicrob Chemother. 2015;70:1960–4.PubMedGoogle Scholar
- Metcalf BJ, Chochua S, Gertz RE Jr, Li Z, Walker H, Tran T, et al. Active Bacterial Core surveillance team. Using whole genome sequencing to identify resistance determinants and predict antimicrobial resistance phenotypes for year 2015 invasive pneumococcal disease isolates recovered in the United States. Clin Microbiol Infect. 2016;22:1002.e1–8. DOIGoogle Scholar
- Beall B, McEllistrem MC, Gertz RE Jr, Wedel S, Boxrud DJ, Gonzalez AL, et al.; Active Bacterial Core Surveillance Team. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J Clin Microbiol. 2006;44:999–1017. DOIPubMedGoogle Scholar
- Beall BW, Gertz RE, Hulkower RL, Whitney CG, Moore MR, Brueggemann AB. Shifting genetic structure of invasive serotype 19A pneumococci in the United States. J Infect Dis. 2011;203:1360–8. DOIPubMedGoogle Scholar
- Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE Jr, Walker H, et al. Penicillin-binding protein transpeptidase signatures for tracking and predicting β-lactam resistance levels in Streptococcus pneumoniae. MBio. 2016;7:e00756–16. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. Twenty-third informational supplement (M100–S22.CLSI). Wayne (PA): The Institute; 2013.
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17:10–2. DOIGoogle Scholar
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. DOIPubMedGoogle Scholar
- Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31:2877–8. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. DOIPubMedGoogle Scholar
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–30. DOIPubMedGoogle Scholar
- Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci U S A. 2006;103:2857–62. DOIPubMedGoogle Scholar
- Coffey TJ, Daniels M, Enright MC, Spratt BG. Serotype 14 variants of the Spanish penicillin-resistant serotype 9V clone of Streptococcus pneumoniae arose by large recombinational replacements of the cpsA-pbp1a region. Microbiology. 1999;145:2023–31. DOIPubMedGoogle Scholar
- Brueggemann AB, Pai R, Crook DW, Beall B. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 2007;3:e168. DOIPubMedGoogle Scholar
- Wyres KL, Lambertsen LM, Croucher NJ, McGee L, von Gottberg A, Liñares J, et al. Pneumococcal capsular switching: a historical perspective. J Infect Dis. 2013;207:439–49. DOIPubMedGoogle Scholar
- Mavroidi A, Aanensen DM, Godoy D, Skovsted IC, Kaltoft MS, Reeves PR, et al. Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J Bacteriol. 2007;189:7841–55. DOIPubMedGoogle Scholar
- Golubchik T, Brueggemann AB, Street T, Gertz RE Jr, Spencer CC, Ho T, et al. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat Genet. 2012;44:352–5. DOIPubMedGoogle Scholar
- Enright MC, Spratt BG. Extensive variation in the ddl gene of penicillin-resistant Streptococcus pneumoniae results from a hitchhiking effect driven by the penicillin-binding protein 2b gene. Mol Biol Evol. 1999;16:1687–95. DOIPubMedGoogle Scholar
- Kauffmann F, Mørch E, Schmith K. On the serology of the pneumococcus-group. J Immunol. 1940;39:397–426.
- Lund E. On the nomenclature of the pneumococcal types. Int J Syst Bacteriol. 1970;20:321–3. DOIGoogle Scholar
- Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol. 1995;33:2759–62.PubMedGoogle Scholar
- Kim L, McGee L, Tomczyk S, Beall B. Biological and epidemiological features of antibiotic-resistant Streptococcus pneumoniae in pre- and post-conjugate vaccine eras: a United States perspective. Clin Microbiol Rev. 2016;29:525–52. DOIPubMedGoogle Scholar
- Olarte L, Kaplan SL, Barson WJ, Romero JR, Lin PL, Tan TQ, et al. Emergence of multidrug-resistant pneumococcal serotype 35B among children in the United States. J Clin Microbiol. 2017;55:724–34. DOIPubMedGoogle Scholar
- Moore MR, Gertz RE Jr, Woodbury RL, Barkocy-Gallagher GA, Schaffner W, Lexau C, et al. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J Infect Dis. 2008;197:1016–27. DOIPubMedGoogle Scholar
- Sobanjo-ter Meulen A, Vesikari T, Malacaman EA, Shapiro SA, Dallas MJ, Hoover PA, et al. Safety, tolerability and immunogenicity of 15-valent pneumococcal conjugate vaccine in toddlers previously vaccinated with 7-valent pneumococcal conjugate vaccine. Pediatr Infect Dis J. 2015;34:186–94. DOIPubMedGoogle Scholar
Page created: May 16, 2017
Page updated: May 16, 2017
Page reviewed: May 16, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.