Volume 23, Number 8—August 2017
Synopsis
Added Value of Next-Generation Sequencing for Multilocus Sequence Typing Analysis of a Pneumocystis jirovecii Pneumonia Outbreak1
Figure 2
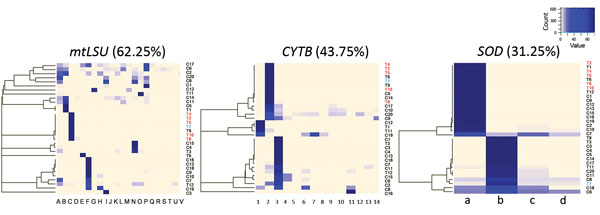
Figure 2. Heatmaps representing distribution for mtLSU, CYTB, and SOD variants among patients infected during a Pneumocystis jirovecii pneumonia outbreak at a university hospital in France, 2014–2015. Patients shown in red correspond to cluster patients sharing the common C2a genotype. Patient shown in blue is the patient having the C2a genotype as minor variant (2%). Percentages shown indicate the level of mixed-strain infections for each locus.
1Preliminary results from this study were presented at the American Society for Microbiology Microbe 2016 Conference, June 16–20, 2016, Boston, Massachusetts, USA.
2Current affiliation: Centre Hospitalier Universitaire Limoges, Limoges, France.
Page created: July 17, 2017
Page updated: July 17, 2017
Page reviewed: July 17, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.