Volume 24, Number 5—May 2018
Dispatch
Second Human Pegivirus in Hepatitis C Virus–Infected and Hepatitis C Virus/HIV-1–Co-infected Persons Who Inject Drugs, China
Abstract
We report the presence of the second human pegivirus (HPgV-2) in Guangdong and Sichuan Provinces in China. The prevalence of HPgV-2 in hepatitis C virus/HIV-1–co-infected persons who inject drugs was 12.9% in Guangdong and 15.9% in Sichuan. This population is at high risk for HPgV-2 infection.
In 2015, the second human pegivirus (HPgV-2) was independently reported by 2 groups in the United States (1,2). Previous reports have indicated that HPgV-2 (also known as HHpgV-1) is a transfusion-transmitted virus and is associated with hepatitis C virus (HCV) infection (1–5). The distribution and prevalence of HPgV-2 infection worldwide are of great importance but remain to be determined. In this study, we demonstrate the existence of HPgV-2 in the southern province of Guangdong and southwestern province of Sichuan in China. We have also identified HCV-infected persons, in particular HCV/HIV-1 co-infected persons who inject drugs (PWID), as populations at high risk for HPgV-2 infection. In addition, our work reveals the difference in the prevalence, distribution, and phylogeny between the first human pegivirus (HPgV; formerly GB virus C or hepatitis G virus) (6,7) and HPgV-2.
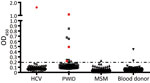
In our initial investigation of HPgV-2, we screened a total of 367 delinked serum or plasma samples from high-risk groups for infection with HCV and HIV-1 and 500 healthy volunteer blood donors from Guangdong Province, China, by using ELISA (2,5), and a nested reverse transcription PCR targeting both the 5′ untranslated region and nonstructural protein 3 regions of HPgV-2 (3,5). We observed a low frequency (0.4%) of HPgV-2 antibody detection and the absence of HPgV-2 viremia in healthy blood donors tested in our study. Out of 86 HCV-infected patients, 1 (1.2%) was positive for both HPgV-2 antibodies and viral RNA (Table 1). Furthermore, we did not detect HPgV-2 RNA in men who have sex with men (MSM), although 1 (0.5%) of the 211 MSM was weakly positive for HPgV-2 antibodies and negative for HPgV-2 RNA (Table 1; Figure 1).
We observed a relatively high prevalence of HPgV-2 infection in HCV/HIV-1 co-infected PWID in Guangdong Province; 12.9% (9/70) were positive for HPgV-2 antibodies and 5.7% (4/70) for HPgV-2 RNA (Table 1). We obtained similar results from 270 PWID from Sichuan Province; 15.9% (43/270) were positive for HPgV-2 antibodies and 3.0% (8/270) for HPgV-2 RNA (Table 1). Using the Fisher exact test, we observed a statistically significant difference between HCV-positive and HCV-negative patients in the prevalence of having HPgV-2 antibodies (6.2% vs. 0; p<0.001) and prevalence of having HPgV-2 RNA (5% vs. 0; p = 0.026). Similarly, we observed a statistically significant difference between HIV-1–positive/HCV-positive patients and HIV-1–positive/HCV-negative patients in the prevalence of having HPgV-2 antibodies (10% vs. 0; p<0.001) and prevalence of having HPgV-2 RNA (4% vs. 0; p = 0.040) (Table 2). These findings indicate a close association between HPgV-2 and HCV infection and synergy between HIV-1 and HCV infection with respect to HPgV-2 infections (5).
Furthermore, we obtained 6 near full-length genome sequences of HPgV-2 by using next-generation sequencing or sequencing of PCR products (5). These strains from China, which included 2 from PWID (IDU31 and SC-LS-01), 2 from HCV-infected patients (HCV-121 and C346), and 2 from HCV-infected blood donors (HCV1241 and HCV1563), exhibited an identity of 93.6%–97.8% at the whole-genome level. Compared with other HPgV-2 strains from the United States and United Kingdom, the nucleotide sequence identity was 93.7%–96.2%. Sequence divergence was greatest at synonymous sites, with ratios of nonsynonymous to synonymous nucleotide substitutions of 0.125–0.150, which are consistent with other reports (1–3). Phylogenetic analysis indicated that HPgV-2 strains from China, the United States, and the United Kingdom clustered together to form a separate branch and fell into group 1 with the closely related pegiviruses from bats and rodents (Figure 2). Other pegiviruses from human, simian, and equine sources formed group 2, in which the variants of HPgV fell into a separate clade. These results illustrate the difference between the 2 human pegiviruses (1,2,8) and the low level of genetic diversity of HPgV-2 strains (1–3).
In contrast to our findings on HPgV-2 infection, we observed a high frequency of HPgV infection across all 3 populations tested (HCV-infected patients, PWID, and MSM) (Tables 1, 2). The percentage of HPgV viremia was 14.0% (14/86) in HCV-infected patients, 19.0% (40/211) in MSM, and 40.0% (28/70) in PWID (Table 1). Among MSM, the prevalence of HPgV RNA was 28.0% (28/100) in those who were infected with HIV-1 alone and 33.3% (4/12) in those who were HIV-1/HCV co-infected (Table 1). For MSM who were negative for both HIV-1 and HCV, 7.9% (7/89) were positive for HPgV RNA (Table 1).
We report the detection of the second human pegivirus, HPgV-2, in HCV-infected (in particular HCV/HIV-1 co-infected) persons in Guangdong and Sichuan Provinces, China (Table 1). Our results and those from previous studies demonstrate that the virus occurs in several geographically distinct regions in the world (1–4,9,10).
HPgV and HPgV-2 are the only known human pegiviruses (8), and comparing their association with HCV and HIV-1 infection is of great interest. Consistent with previous reports, we found that the prevalence of HPgV viremia was 7.9% in HCV and HIV-1–negative MSM and 33.3%–40% in HCV/HIV-1 co-infected MSM and PWID (Table 1). In contrast, only 0.5% of MSM and 0.4% of healthy blood donors were positive for HPgV-2 antibodies, but all were negative for HPgV-2 RNA (Table 1). These results indicate that HPgV-2 infection might be much less frequent than HPgV infection, possibly because of its low transmissibility or high clearance rate (2–4). The dramatic difference of distribution and prevalence between HPgV and HPgV-2 infections in different populations provides a clue for investigation of disease association with HPgV-2. HPgV does not cause human diseases (11) and can inhibit HIV-1 replication as well as prolong survival of HIV-1–infected and Ebola virus–infected patients (12–14). However, possible pathogenicity and disease association of HPgV-2 remain to be elucidated.
The high-risk populations susceptible to HPgV-2 infection include HCV-infected patients and, in particular, HCV/HIV-1 co-infected PWID. Most (93.3%) of HPgV-2 infected patients were also co-infected with HCV (1–4). Notably, the relatively high frequency of HPgV-2 RNA detection was observed in HCV/HIV-1 co-infected PWID in Guangdong (5.7%) and Sichuan (3.0%) Provinces of China (Table 1) and in the United States (10.9%) (9). In contrast, a somewhat lower percentage (1.7%) of HCV-positive PWID in the United Kingdom were reported to be HPgV-2 RNA positive, whereas none of the 30 HIV-1 singly infected and 36 HCV/HIV-1 co-infected PWID were positive for HPgV-2 RNA (3). These discordant results warrant more studies in different countries to address the association between HPgV-2 and HCV/HIV-1 co-infection.
Our findings are subject to 2 limitations. First, because a limited number of samples from only 2 provinces of China were tested, the results might not represent overall prevalence of HPgV-2 infection throughout all of China. Second, this study was a cross-sectional rather than a longitudinal study, therefore, the proportions of persistent infection and natural history of HPgV-2 infection remain to be determined.
Future studies should address several questions: whether the close association between HPgV-2 and HCV infection represents a biologic dependence of these 2 viruses; how HCV/HIV-1 co-infection facilitates HPgV-2 infection; and whether HCV or HIV-1 viral proteins enhance the transmissibility or infectivity of HPgV-2. In addition, because the rarity of HPgV-2 detection in MSM could be a result of the low frequency of HCV or HIV-1 infection or the transmission route of HPgV-2, further research should aim to determine if HPgV-2 is more like a transfusion-transmitted virus rather than a sexually transmitted virus.
Dr. Wang is a postdoctoral fellow at the School of Public Health of Southern Medical University in Guangzhou. Her research interests include identification and diagnosis of viral pathogens and investigation of viral pathogenesis.
Acknowledgment
This work was supported by the Bureau of Science and Information Technology of Guangzhou Municipality (grant nos. 201604020011, 2014Y2-00550, and 201704020219) and the Beijing Municipal Science and Technology Project (grant no. D141100000314001).
References
- Kapoor A, Kumar A, Simmonds P, Bhuva N, Singh Chauhan L, Lee B, et al. Virome analysis of transfusion recipients reveals a novel human virus that shares genomic features with hepaciviruses and pegiviruses. MBio. 2015;6:e01466–15. DOIPubMedGoogle Scholar
- Berg MG, Lee D, Coller K, Frankel M, Aronsohn A, Cheng K, et al. Discovery of a novel human pegivirus in blood associated with hepatitis C virus co-infection. PLoS Pathog. 2015;11:e1005325. DOIPubMedGoogle Scholar
- Bonsall D, Gregory WF, Ip CL, Donfield S, Iles J, Ansari MA, et al. Evaluation of viremia frequencies of a novel human pegivirus by using bioinformatic screening and PCR. Emerg Infect Dis. 2016;22:671–8. DOIPubMedGoogle Scholar
- Coller KE, Berg MG, Frankel M, Forberg K, Surani R, Chiu CY, et al. Antibodies to the novel human pegivirus 2 are associated with active and resolved infections. J Clin Microbiol. 2016;54:2023–30. DOIPubMedGoogle Scholar
- Wang H, Wan Z, Xu R, Guan Y, Zhu N, Li J, et al. A novel human pegivirus, HPgV-2 (HHpgV-1), is tightly associated with hepatitis C virus (HCV) infection and HCV/human immunodeficiency virus type 1 coinfection. Clin Infect Dis. 2018;66:29–35. DOIPubMedGoogle Scholar
- Simons JN, Leary TP, Dawson GJ, Pilot-Matias TJ, Muerhoff AS, Schlauder GG, et al. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–9. DOIPubMedGoogle Scholar
- Linnen J, Wages J Jr, Zhang-Keck ZY, Fry KE, Krawczynski KZ, Alter H, et al. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–8. DOIPubMedGoogle Scholar
- Smith DB, Becher P, Bukh J, Gould EA, Meyers G, Monath T, et al. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J Gen Virol. 2016;97:2894–907. DOIPubMedGoogle Scholar
- Kandathil AJ, Breitwieser FP, Sachithanandham J, Robinson M, Mehta SH, Timp W, et al. Presence of human hepegivirus-1 in a cohort of people who inject drugs. Ann Intern Med. 2017;167:1–7. DOIPubMedGoogle Scholar
- Frankel M, Forberg K, Coller KE, Berg MG, Hackett J Jr, Cloherty G, et al. Development of a high-throughput multiplexed real time RT-PCR assay for detection of human pegivirus 1 and 2. J Virol Methods. 2017;241:34–40. DOIPubMedGoogle Scholar
- Mohr EL, Stapleton JT. GB virus type C interactions with HIV: the role of envelope glycoproteins. J Viral Hepat. 2009;16:757–68. DOIPubMedGoogle Scholar
- Vahidnia F, Petersen M, Stapleton JT, Rutherford GW, Busch M, Custer B. Acquisition of GB virus type C and lower mortality in patients with advanced HIV disease. Clin Infect Dis. 2012;55:1012–9. DOIPubMedGoogle Scholar
- Zhang W, Chaloner K, Tillmann HL, Williams CF, Stapleton JT. Effect of early and late GB virus C viraemia on survival of HIV-infected individuals: a meta-analysis. HIV Med. 2006;7:173–80. DOIPubMedGoogle Scholar
- Lauck M, Bailey AL, Andersen KG, Goldberg TL, Sabeti PC, O’Connor DH. GB virus C coinfections in west African Ebola patients. J Virol. 2015;89:2425–9. DOIPubMedGoogle Scholar
Figures
Tables
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 24, Number 5—May 2018
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Shixing Tang, Southern Medical University, School of Public Health, 1838 N Guangzhou Ave, Guangzhou, 510515, China
Top