Volume 25, Number 6—June 2019
Dispatch
New Delhi Metallo-β-Lactamase 5–Producing Klebsiella pneumoniae Sequence Type 258, Southwest China, 2017
Cite This Article
Citation for Media
Abstract
We isolated a New Delhi metallo-β-lactamase 5 (NDM-5)–producing Klebsiella pneumoniae sequence type (ST) 258 strain in southwest China during 2017. The blaNDM-5 gene was acquired by horizontal plasmid transfer from NDM-5–producing Escherichia coli. We identified genomic characteristics in ST258 strains that differed from those of global K. pneumoniae carbapenemase–producing strains.
Carbapenem-resistant Klebsiella pneumoniae has emerged as one of the major multidrug-resistant bacterial pathogens worldwide. The international spread of this pathogen has been linked to a few high-risk clone group (CG) strains, including CG258, CG14/15, CG17/20, CG43, CG147, and CG307 (1), of which CG258 is the most widely spread. Within CG258, sequence type (ST) 258 predominates in North America and Europe, but ST11 is the primary carbapenem-resistant K. pneumoniae clone in eastern Asia, especially in China (2–4).
In recent years, K. pneumoniae ST11 strains have been increasingly identified in North America. In contrast, ST258 strains are still rarely reported in China (5). Unlike other high-risk clones, such as ST11, that harbor different types of carbapenemases, carbapenem-resistant K. pneumoniae ST258 are almost exclusively associated with K. pneumoniae carbapenemase (KPC) (6). We report identification of a New Delhi metallo-β-lactamase 5 (NDM-5)–producing K. pneumoniae ST258 strain from Chengdu in southwest China.
In June 2017, a K. pneumoniae strain (Kp2588) that has an extended-spectrum β-lactamase phenotype was isolated from a blood culture of a male patient in a large tertiary care hospital in Chengdu in southwest China. Two days later, a carbapenem-resistant Escherichia coli strain (Ec2551) was isolated from a urine culture, and 1 week later, a carbapenem-resistant K. pneumoniae strain (Kp2573) was isolated from another urine culture, both from the same patient (Appendix).
PCR testing and Sanger sequencing showed that Ec2551 and Kp2573 both harbored the New Delhi metallo-β-lactamase gene blaNDM-5. Multilocus sequence typing further showed that Ec2551 is an ST48 strain, and Kp2588 and Kp2573 belong to the high-risk clone ST258.
Susceptibility testing showed that the initial Kp2588 blood isolate was resistant to most cephalosporins and aztreonam but susceptible to cefepime, carbapenems, amoxicillin/clavulanic acid, piperacillin/tazobactam, amikacin, and tigecycline (Table). In contrast, the urinary isolates Ec2551 and Kp2573 were resistant to all β-lactams and β-lactam/β-lactamase inhibitors, including the 4 carbapenems tested in this study (Table).
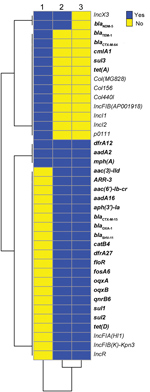
We conducted next-generation sequencing (NGS; Illumina Hiseq, https://www.illumina.com) for all 3 isolates and deposited data in the National Center for Biotechnology (NCBI) Bioproject PRJNA354234 (https://www.ncbi.nlm.nih.gov/bioproject) GenBank database (accession nos. RXHE00000000, RXHG00000000 and RXHF00000000, respectively). Analysis of resistance genes and plasmid sequences showed that the 2 ST258 K. pneumoniae isolates carried similar resistance and plasmid replicon genes, except that Kp2573 harbored blaNDM-5 and an IncX3 plasmid replicon (Figure 1). E. coli strain Ec2551 had the same blaNDM-5 and IncX3 replicon genes as Kp2573, but also had a diverse array of other resistance and plasmid replicon genes in comparison with the 2 K. pneumoniae isolates (Figure 2).
We then transferred blaNDM-5–harboring plasmids from Ec2551 and Kp2573 to recipient strain E. coli J53AZR by conjugation, followed by plasmid NGS as described. Plasmids sequence analysis showed that Ec2551 and Kp2573 carry an identical blaNDM-5–harboring IncX3 plasmid. The plasmid is 46,161 bp and is identical to several blaNDM-5–harboring plasmids identified in China and other countries (e.g., GenBank accession nos. CP032424, MF679143, MG591703, CP028705, and CP024820). Core single-nucleotide polymorphism (SNP) analysis showed that Kp2588 differed from Kp2573 by 1 SNP. These results strongly suggest that carbapenem-resistant Kp2573 might have evolved from the carbapenem-susceptible blood isolate Kp2588 by acquisition of a blaNDM-5–harboring IncX3 plasmid from the same patient.
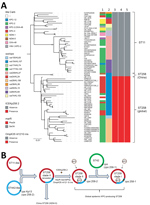
To date, carbapenemases other than KPC have rarely been described in clinical K. pneumoniae ST258 strains (7,8). In this study, we had the opportunity to investigate the phylogenetic relationship between NDM-5–producing ST258 (designated as the China ST258 clone) and other global KPC-producing ST258 strains (Figure 2, panel A). Phylogenetic analysis of core SNPs from the 2 China ST258 genomes and 76 completely closed ST258 (n = 39) and ST11 (n = 36) genomes from NCBI showed that Kp2573/Kp2588 belongs to a separate clade distinct from global ST258 and ST11 strains (Figure 2, panel A). Kp2573/Kp2588 differs from global ST258 genomes by an average of 88 core SNPs (range 54–135) and from ST11 strains by an average of 109 core SNPs (range 79–132).
Results indicated that Kp2573/Kp2588 and the global ST258 strains both evolved from a common ancestor. A different core SNP phylogenetic analysis of Kp2573/Kp2588 with 824 ST258 genomes (including both draft assemblies and completely closed genomes) from NCBI showed similar results, suggesting that Kp2573/Kp2588 has unique genetic characteristics in comparison with other global ST258 strains.
Recent phylogenetic analysis showed that global ST258 strains have unique genetic characteristics that might contribute to their epidemic spread. For example, global ST258 strains carry a unique integrative and conjugative element (ICE) known as ICEKp258.2 (3). This chromosomal element is conserved among nearly all ST258 strains, harboring a type IV pilus gene cluster and a type III restriction-modification system, which might contribute to plasmid acquisition or plasmid–host specificity (9). However, ICEKp258.2 is absent from genomes of Kp2573/Kp2588 (Figure 2, panel A). In addition, global ST258 strains have a common Ser34Phe amino acid substitution in the homodimerization region of MarR, a transcriptional regulator protein of the multiple antimicrobial drug resistance repressor family (10). It is hypothesized that this substitution might affect the overall metabolic activity in ST258 (10). Kp2573/Kp2588 has the ancestral MarR Ser34 genotype, similar to ST11 strains, but all global ST258 strains have the Phe34 genotype. Furthermore, all global ST258 strains contain a frame-shift mutation that results in a premature stop codon at amino acid position 89 in outer membrane protein OmpK35 because of a guanine insertion at nt position 121. In contrast, Kp2573/Kp2588 do not have the nt121G insertion in the ompK35 gene but have an IS1 insertion at nt position 28. These genetic changes further suggest that the 2 China ST258 strains have distinct evolutionary paths compared with those of global ST258 strains.
These observations have also updated our previous hypothesis regarding the molecular evolution of ST258 (Figure 2, panel B) (9). Our initial study suggested that the prototypical ST258 strain arose as a consequence of recombination of large segments of the ST11 and ST442 genomes, such that it has the same wzi154/KL107 (previously named cps-2/clade II) capsular type as the parental ST442 strain (9). Additional genotypic changes, including acquisition of ICEKp258.2, the MarR Ser34Phe substitution, and the ompK35 nt121G insertion, as well as the acquisition of KPC plasmids in ST258 clade II strains, define the global KPC-producing carbapenem-resistant K. pneumoniae ST258 clone.
A second molecular event, generated by replacement of the ST258 clade II cps region with the corresponding region from ST42, subsequently created the equally prevalent ST258 clade I clone (9). Both clades I and II ST258 strains have shown the ability to host diverse KPC plasmids, which might in part contribute to the spread of global ST258 strains. In contrast, the China ST258 strains appear to have evolved separately, recently acquiring the blaNDM5–harboring plasmid (Figure 2, panel B), highlighting the continuing evolution of K. pneumoniae ST258 strains.
We report the genotype of an NDM-5–producing K. pneumoniae ST258 strain isolated in southwest China. The blaNDM-5 gene was likely acquired by a carbapenem-susceptible ST258 strain from an NDM-5–producing E. coli strain in vivo in the same patient as a result of horizontal transfer of a blaNDM-5–harboring IncX3 plasmid. Genomic comparison of the ST258 strain from China with other carbapenem-resistant Klebsiella pneumoniae ST258 strains indicated that they both evolved from a common ancestral strain but with distinct genetic characteristics suggestive of separate evolutionary histories. Further genomic comparisons between ST258 strains from China and their global ST258 counterparts will help elucidate the molecular mechanisms underscoring the spread of this high-risk clone.
Dr. Xin Zhang is a clinical microbiologist at the Sichuan Academy of Medical Science, Sichuan, China. Her research interests include the clinical epidemiology of antimicrobial drug resistance and infectious disease.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (81572032); the Six Talent Peaks Project in Jiangsu Province (2016-WSN-112); the Key Research and Development Project of Jiangsu Provincial Science and Technology Department (BE2017654); Gusu Key Health Talent of Suzhou; the Jiangsu Youth Medical Talents Program (QN-866, 867); the Science and Technology Program of Suzhou (SZS201715 and SYS201619); and the National Institutes of Health (R01AI090155, R21AI117338, and P30CA008748).
References
- Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant cones. Trends Microbiol. 2016;24:944–56. DOIPubMedGoogle Scholar
- Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013;4:48. DOIPubMedGoogle Scholar
- Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–96. DOIPubMedGoogle Scholar
- Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. DOIPubMedGoogle Scholar
- Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JD. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob Agents Chemother. 2013;57:130–6. DOIPubMedGoogle Scholar
- Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41:252–75. DOIPubMedGoogle Scholar
- Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28:565–91. DOIPubMedGoogle Scholar
- Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59:5873–84. DOIPubMedGoogle Scholar
- Chen L, Mathema B, Pitout JD, DeLeo FR, Kreiswirth BN. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. MBio. 2014;5:e01355–14. DOIPubMedGoogle Scholar
- Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, Lemmer D, et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One. 2015;10:
e0133727 . DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: May 03, 2019
1These authors contributed equally to this article.
Table of Contents – Volume 25, Number 6—June 2019
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Hong Du, Department of Clinical Laboratory, the Second Affiliated Hospital of Soochow University, 1055 Sanxiang Rd, Suzhou, Jiangsu, 215004, China
Top