Volume 26, Number 3—March 2020
Research
Genomic and Phenotypic Variability in Neisseria gonorrhoeae Antimicrobial Susceptibility, England
Figure 1
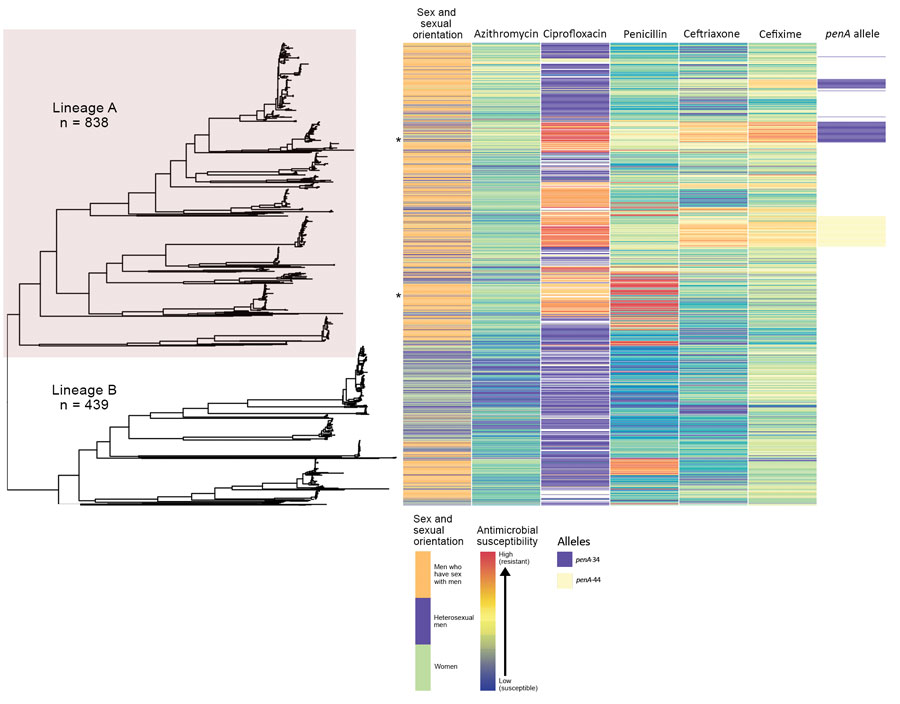
Figure 1. Phylogeny and antimicrobial susceptibility of Neisseria gonorrhoeae isolates from England, 2013–2016. Maximum-likelihood phylogeny with recombination events removed of all N. gonorrhoeae isolates annotated with gender and sexual orientation, antimicrobial susceptibility phenotype, and penA genotype. Asterisks represent location in tree of isolates with high-level azithromycin resistance (MIC >256 mg/L). Heterosexual men were those who reported sex with women exclusively.
Page created: February 20, 2020
Page updated: February 20, 2020
Page reviewed: February 20, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.