Volume 26, Number 3—March 2020
Research
Genomic and Phenotypic Variability in Neisseria gonorrhoeae Antimicrobial Susceptibility, England
Cite This Article
Citation for Media
Abstract
Antimicrobial resistance (AMR) in Neisseria gonorrhoeae is a global concern. Phylogenetic analyses resolve uncertainties regarding genetic relatedness of isolates with identical phenotypes and inform whether AMR is due to new mutations and clonal expansion or separate introductions by importation. We sequenced 1,277 isolates with associated epidemiologic and antimicrobial susceptibility data collected during 2013–2016 to investigate N. gonorrhoeae genomic variability in England. Comparing genetic markers and phenotypes for AMR, we identified 2 N. gonorrhoeae lineages with different antimicrobial susceptibility profiles and 3 clusters with elevated MICs for ceftriaxone, varying mutations in the penA allele, and different epidemiologic characteristics. Our results indicate N. gonorrhoeae with reduced antimicrobial susceptibility emerged independently and multiple times in different sexual networks in England, through new mutation or recombination events and by importation. Monitoring and control for AMR in N. gonorrhoeae should cover the entire population affected, rather than focusing on specific risk groups or locations.
Antimicrobial resistance (AMR) in Neisseria gonorrhoeae is a global concern and affects all classes of antimicrobial drugs used for treatment. Penicillin, ciprofloxacin, and cefixime were the recommended first-line antimicrobial drug therapies until AMR prevalence breached the World Health Organization (WHO) recommended threshold of >5% of local isolates demonstrating resistance; at that point, ceftriaxone became the preferred antimicrobial drug treatment (1). However, ceftriaxone resistance has been reported in many countries and frequently in East and Southeast Asia, probably because of poor antimicrobial stewardship (2). Ceftriaxone resistance has been linked to mutations in the penA gene, which has been reported in several continents, including North America and Europe (3–5).
To clarify the spread of AMR in N. gonorrhoeae and the population groups most at risk, surveillance programs and research studies often link phenotypic susceptibility data with data on the epidemiologic characteristics of cases (6,7). However, these analyses are limited because isolates with identical phenotypes might not be genetically related. Consequently, determining the extent to which AMR transmission is due to clonal dissemination or separate introductions is challenging and these data are essential to guide the public health response.
Combining phenotypic and genomic data can help resolve uncertainties. Whole-genome sequencing (WGS) enables investigation of genetic determinants for AMR and how these are distributed in the pathogen population (4,5). WGS also can contribute evidence toward the development of rapid antimicrobial susceptibility tests to improve treatment decisions (8,9). However, few N. gonorrhoeae WGS studies have been conducted in England, and none include representative geographic coverage over time (10–13).
We investigated the genomic and phenotypic variability in N. gonorrhoeae antimicrobial susceptibility in England. We described the epidemiologic characteristics of genetically distinct clusters of infection with reduced susceptibility to cefixime, ceftriaxone, and azithromycin and resistance to ciprofloxacin and penicillin. We focused on N. gonorrhoeae with mutations in the penA allele, which contribute to reduced susceptibility to ceftriaxone. In addition, we assessed the genetic similarity of N. gonorrhoeae in England, Europe, and the United States to determine the extent to which international travel might influence the spread of AMR in N. gonorrhoeae.
Isolate Selection
We selected N. gonorrhoeae isolates from the archives of the Gonococcal Resistance to Antimicrobials Surveillance Programme (GRASP), a sentinel program implemented by Public Health England (PHE) in 2000. GRASP is designed to represent the gonococcal population in England (14) and includes clinical, sociodemographic, and behavioral data collected through the GUMCAD STI Surveillance System (https://www.gov.uk/guidance/gumcad-sti-surveillance-system) and directly from clinics. During a 3-month period each year, GRASP tests for antimicrobial susceptibility in consecutive isolates from all culture-positive N. gonorrhoeae cases identified in 26 sexual health clinics in England and Wales (15). GRASP collects ≈1,200–2,500 isolates annually for antimicrobial susceptibility testing (16).
We selected isolates collected from 5 GRASP clinics, 2 in London and 3 in other geographically distinct areas in England: Birmingham, Bristol, and Liverpool. We chose these locations to obtain isolates from cases representing a broad range of sociodemographic and behavioral characteristics, including sex, sexual orientation, age, ethnicity, and HIV status (Appendix 1 Table 1). We sequenced all isolates collected during 2013–2016 by the 5 clinics and stored in the GRASP archive. We chose the most recent years of GRASP data to investigate prevailing trends and patterns. We did not include isolates from a 2015 outbreak of high-level azithromycin-resistant N. gonorrhoeae in the United Kingdom in this sampling frame because the isolates did not meet the eligibility criteria of our study.
Ethics Considerations
PHE has permission to process confidential patient data obtained by GRASP under Regulation 3 (Control of Patient Information) of the Health Service Regulations 2002. Information governance advice and ethics approval for this study were granted by the PHE Research Ethics and Governance Group.
Antimicrobial Susceptibility Testing
GRASP tests isolates for antimicrobial susceptibility by using agar dilution methods, records MICs for antimicrobial drugs, and defines AMR by using European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints. For our study, we used data on MICs for ceftriaxone, azithromycin, cefixime, penicillin, and ciprofloxacin (17). In GRASP, epidemiologic data were linked to MICs for antimicrobial drugs for each isolate phenotype.
Isolation and WGS
We retrieved selected isolates from the GRASP archive by culturing on nonselective gonococcus agar (Difco BBL GC II Agar Base [Becton, Dickinson and Company, https://www.bd.com] plus 1% Vitox [Oxoid, http://www.oxoid.com]). We extracted DNA from a subculture of a single colony of each isolate by using the automated QIAsymphony DNA Mini Kit (QIAGEN, https://www.qiagen.com). WGS was conducted at the Wellcome Sanger Institute (Cambridge, UK) by using the HiSeq X Ten system (Illumina, https://www.illumina.com) and processed in the routine Sanger WGS data management pipeline (Appendix 1).
Data Sources from Europe and the United States
We compared the study sample with published WGS and associated metadata for N. gonorrhoeae isolates from international studies. The collection from Europe (European Nucleotide Archive (ENA; accession no. PRJEB9227) contained 1,054 isolates from the European Gonococcal Antimicrobial Surveillance Programme (Euro-GASP) (5). Isolates were collected in 2013 from 20 countries and included 106 isolates from England. We excluded 21% (22/106) that were duplicates of isolates in the study sample, leaving 948 isolates from Europe. The metadata for isolates from Europe included reporting country, antimicrobial susceptibility profile, MICs for ceftriaxone and cefixime, and the presence of the penA-34 allele. We grouped MICs to match the categories used in GRASP.
Isolates from the United States were from 2 previous studies investigating the association between phenotype and genotype for AMR in N. gonorrhoeae (4,9). The collection from the United States contained 1,114 isolates collected during 2000–2013 (ENA accession nos. PRJEB2999 and PRJEB7904). The metadata for the isolates from the United States included antimicrobial susceptibility profiles; 270 had reduced susceptibility to cephalosporin (MIC >0.25 mg/L for cefixime or MIC >0.125 mg/L for ceftriaxone); 294 had reduced susceptibility to azithromycin (MIC >2 mg/L); and 594 were ciprofloxacin-resistant (MIC >1 mg/L). Metadata also included sexual orientation of case-patients and the presence of the penA-34 allele. We grouped MICs to match the categories used in GRASP.
Phylogenetic Analysis
We created phylogenetic trees and removed genetic recombination events by using default settings in Gubbins version 2.4.0 (18), including 5 iterations and >3 base substitutions to identify a recombination event, and the RAxML (Geneious, https://www.geneious.com) or FastTree (19) tree building option (Appendix 1). We created 3 phylogenetic trees: isolates from England only, isolates from England and other countries in Europe, and isolates from England and the United States.
We identified known genetic markers of AMR, including mutations in the penA allele, by using ARIBA (20). We compared MICs to the genetic markers by using the ARIBA micplot module.
We identified the genotype of isolates in large and distinct clusters of N. gonorrhoeae with elevated MICs for ceftriaxone (MIC ≥0.015 mg/L) and cefixime (MIC ≥0.03 mg/L) from the phylogenetic trees. We compared the epidemiologic characteristics of cases in the clusters by using the χ2 or Fisher exact test.
Statistical Analysis
We used univariate and multivariable analyses to assess differences in the epidemiologic characteristics and antimicrobial susceptibility of isolates between lineages identified in the phylogenetic tree. We analyzed the following explanatory variables: year and location the isolate was collected; case-patient information, including gender, sexual orientation, age, ethnicity, country of birth, whether they had a symptomatic N. gonorrhoeae infection or previous sexually transmitted infection (STI), HIV status, and the number of sexual partners they had in the United Kingdom or through travel-associated sexual partnerships <3 months before diagnosis; and isolate susceptibility data, including reduced susceptibility to ceftriaxone (MIC >0.015 mg/L), cefixime (MIC >0.03 mg/L), or azithromycin (MIC >0.25 mg/L); or resistance to penicillin (MIC >1 mg/L or β-lactamase positive) or ciprofloxacin (MIC >0.06 mg/L). We used elevated MIC thresholds for ceftriaxone, cefixime, and azithromycin to provide a robust sample size for regression analysis.
We also explored the relationship between travel-associated sexual partnerships and reduced susceptibility to antimicrobial drugs by conducting univariate and multivariable analyses with reduced susceptibility or resistance as the outcome and travel-associated sexual partnerships as the primary explanatory variable. We considered CI of the odds ratio (OR) >1.0 and p<0.05 by χ2 test as statistically significant.
We developed multivariable logistic regression models by using a forward approach and including only statistically significant variables associated with the outcome in the univariate model to control for possible confounding between variables. We used the likelihood ratio test to determine which explanatory variables should remain in the multivariable model by using p<0.05 as the threshold of statistical significance.
Sample Description
Of the eligible isolates, we successfully sequenced 91% (1,277/1,407); the bacteria of the remaining 130 isolates were no longer viable for DNA extraction. For all antimicrobial drugs tested, the MIC distributions of the study isolates were similar to those of all GRASP isolates (Appendix 1 Table 2). We found that 3.6% of isolates were resistant to azithromycin (MIC >0.5 mg/L) and 2 isolates were highly resistant (MIC >256 mg/L); 0.6% were resistant to cefixime (MIC >0.125 mg/L), 36.3% to ciprofloxacin (MIC >0.06 mg/L), 16.6% to penicillin (MIC >1 mg/L), and none to ceftriaxone (MIC >0.125 mg/L). The MIC distribution of the isolates not sequenced was similar to the distribution of the sequenced isolates. Most (69%; 881/1,277) isolates were from genital infections, 23.4% (299/1,277) were from rectal infections, and 6.3% (80/1,277) were from pharyngeal infections. Overall, we identified 226 different sequence types (STs) by using N. gonorrhoeae multiantigen sequence typing (NG-MAST) (Appendix 1). We deposited novel sequences extracted for this study into ENA (accession no. ERP022090) and provide metadata (Appendix 2).
N. gonorrhoeae Lineages Circulating in England
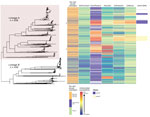
We noted 2 distinct lineages in the phylogenetic tree (Figure 1). Compared with lineage B, lineage A was more likely to contain isolates from clinics in London than those outside of London (outside London:in London adjusted odds ratio [aOR] 1.74, 95% CI 1.27–2.67; p = 0.001). Lineage A also contained more isolates from persons >35 years of age than persons <24 years of age (aOR 1.68, 95% CI 1.16–2.40; p = 0.006). Asian ethnicity also was associated more frequently with isolates from lineage A compared with white ethnicity (aOR 1.86, 95% CI 1.01–3.45; p = 0.048) (Table 1). Lineage A was less likely to contain isolates from women (aOR 0.14, 95% CI 0.09–0.22; p<0.001) or men who reported having sex with women exclusively (MSW; aOR 0.33, 95% CI 0.23–0.47; p<0.001) compared with MSM. This lineage also was less likely to contain isolates from persons reporting black Caribbean ethnicity compared with persons reporting white ethnicity (aOR 0.49, 95% CI 0.32–0.76; p = 0.001). Lineage A was more likely to contain isolates from persons who reported a travel-associated sexual partnership compared with isolates from lineage B (crude odds ratio [cOR] 1.96, 95% CI 1.20–3.21; p = 0.006), but this association did not persist in the multivariable model (aOR 1.66, 95% CI 0.94–2.91; p = 0.078 adjusting for location, age, sex, sexual orientation, and ethnicity). However, isolates from persons who had a recent travel-associated sexual partnership were more likely to be resistant to ciprofloxacin (aOR 1.83, 95% CI 1.13–2.96; p = 0.015, adjusting for location, age, sex, sexual orientation, and ethnicity). We saw no statistically significant association between recent travel-associated sexual partnerships and N. gonorrhoeae with reduced susceptibility to ceftriaxone, cefixime, or azithromycin, or resistance to penicillin.
Isolates with reduced susceptibility to ceftriaxone, cefixime, or azithromycin or resistance to ciprofloxacin and penicillin were dispersed throughout the phylogenetic tree (Figure 1). However, compared with lineage B, isolates in lineage A were more likely to have higher MICs for ceftriaxone (aOR 15.4, 95% CI 8.50–27.8; p<0.001), cefixime (aOR 3.97, 95% CI 2.76–5.76; p<0.001), azithromycin (aOR 7.5, 95% CI 5.37–10.5; p<0.001), and penicillin (aOR 18.2, 95% CI 11.4–29.2; p<0.001) (Table 2).
Distribution of penA Alleles across the Phylogeny
Overall, we identified 32 different known mutations in 8 genes associated with resistance to ceftriaxone, cefixime, azithromycin, ciprofloxacin, or penicillin. For all antimicrobial drugs, we noted isolates with the same combination of genotypic markers of resistance but differing phenotypic MICs (Appendix 1 Figures 1–5).
The larger, distinct clusters with elevated MICs for ceftriaxone and cefixime contained the penA-34 allele and the penA-44 allele (Figure 1). All isolates with the penA-34 allele (n = 86) had a MIC of >0.015 mg/L for cefixime and 67 had a MIC of >0.015 mg/L for ceftriaxone. Most (81/84; 96%) isolates with the penA-44 allele had a MIC of >0.015 mg/L for cefixime, a MIC of >0.015 mg/L for ceftriaxone (83/84; 98%), or both (81/84; 96%).
The 2 largest clusters with the penA-34 allele (cluster 1, n = 57; cluster 2, n = 26) were genetically distinct from each other and isolates in the 2 groups had statistically significant differences by year, clinic, sexual orientation, and HIV status (Table 3). Most (81%; 21/26) isolates in cluster 2 were from London in 2014–2015, and most (67%; 38/57) in cluster 1 were from outside London but distributed across all 4 years of the study. Most (96%; 25/26) isolates in cluster 2 were from MSM, whereas cluster 1 was more mixed and composed of isolates from women (21%; 12/57), MSW (37%; 21/57), and MSM (42%; 24/57). Cluster 2 had a higher percentage of persons living with HIV (35%; 9/26) than did cluster 1 (7%; 4/57). Most (82%; 69/84) isolates with the penA-44 allele were from MSM, persisted over all 4 years of the study, and were found both inside and outside of London. The characteristics of isolates with the penA-44 allele were more similar to the characteristics of isolates in cluster 2 of the penA-34 group than to isolates in cluster 1 (Appendix 1 Table 3).
Comparison of Isolates from England, Europe, and the United States
Isolates from England were genetically interspersed with isolates from other countries in Europe (Figure 2) or the United States (Figure 3), although some large clades of isolates came only from England or the United States. Isolates in cluster 1 with the penA-34 allele in England were clustered with isolates from Europe and the United States (Figures 2–3). Isolates with the penA-34 allele in cluster 2 from England that were only found after 2013 were not related genetically to isolates from the United States or from other countries in Europe.
We conducted a large study on genomic variability of antimicrobial susceptibility in N. gonorrhoeae in England. We sampled isolates from geographically dispersed clinics in England, and our data likely represent patterns at a national level. Our data suggest that N. gonorrhoeae with reduced susceptibility to antimicrobial drugs, including ceftriaxone, has emerged in England through novel mutation and recombination events, repeated introduction from overseas, clonal expansion, or a combination of these.
We observed 3 distinct clusters with 2 different penA alleles and reduced susceptibility to ceftriaxone and cefixime and found patients in each cluster with differing epidemiologic characteristics. The genetic similarity of isolates from England, Europe, and the United States is consistent with global dissemination of N. gonorrhoeae concerning genotypic and phenotypic features. Our data highlight the potential influence of travel-associated sexual partnerships in AMR transmission.
As seen in other N. gonorrhoeae studies, the high frequency of DNA recombination requires computational strategies to use single-nucleotide polymorphism differences arising through mutation rather than recombination. We identified and removed recombinant DNA, but some likely remained, which might lead to incorrect inferences about relatedness for some isolates.
We found 2 distinct lineages of N. gonorrhoeae with different antimicrobial susceptibility profiles circulating in England. The larger lineage contained isolates with elevated MICs or resistance to all 5 antimicrobial drugs tested, consistent with findings from recent studies in Europe and globally (5,21). The authors of those studies hypothesized that differing susceptibility profiles of the 2 lineages were associated with different sexual orientation networks, but neither study had complete data on sexual orientation to support the hypothesis. Our study includes sexual orientation data for 99% of cases. Our findings strongly support the hypothesis that MSM are more frequently infected with N. gonorrhoeae strains with reduced susceptibility to antimicrobials, whereas MSW and women are more frequently infected with the more susceptible lineage. Nevertheless, many MSW were infected with N. gonorrhoeae with reduced susceptibility to antimicrobial drugs and MSM were infected by antimicrobial susceptible N. gonorrhoeae, and we noted intralineage variation by sexual orientation. MSM have more bacterial STI diagnoses and greater exposure to antimicrobial drugs, thereby increasing selection pressures for AMR, a hypothesis supported by mathematical models (21–24). Resistant strains also might persist in the absence of selective pressure because the organism’s biologic fitness is unaffected or compensatory mutations mitigate a deleterious effect (25–27).
By combining WGS, epidemiologic, and phenotypic data, we found that reduced susceptibility to ceftriaxone and cefixime emerged repeatedly in separate sexual networks in England. Without WGS data, we would have grouped all penA-34 samples from MSM together. Likewise, if we restricted sequencing to the penA gene, we would not have identified unique clusters with the same penA-34 allele.
The large group of isolates in England with the penA-34 allele clustered with isolates from Europe and the United States that had the same allele. Some of the penA-34 allele isolates belonged to the NG-MAST 1407 lineage, a widely disseminated clone associated with elevated MICs for ceftriaxone and cefixime and the catalyst for changing national treatment guidelines in the United Kingdom from cefixime as first-line therapy to ceftriaxone in 2011 (6,22). Therefore, the larger penA-34 group probably represents clonal spread of a previously identified endemic strain of N. gonorrhoeae; the smaller penA-34 group represents a new strain emerging in a different sexual network largely comprising MSM in London with a history of STIs, including HIV. Consequently, restricting public health resources that measure, prevent, and control AMR in N. gonorrhoeae to specific risk groups or geographic locations could be ineffective because AMR appears to emerge independently in different sexual networks and locations.
We found some evidence for importation of AMR. Isolates from persons who recently had a travel-associated sexual partnership were more likely to be infected with N. gonorrhoeae that was resistant to ciprofloxacin. Although de novo development of high-level resistance to azithromycin in the United Kingdom has been described, some studies have concluded that importation events probably initiate AMR spread in countries with low population prevalence, such as England (11,27,28). The success of antimicrobial stewardship policies and compliance with treatment guidelines that aim to curtail AMR in the endemic gonococcal population in England could be undermined by the importation and subsequent spread of resistant isolates. These data support the importance of promoting STI prevention messages and testing to international travelers, particularly those visiting countries where AMR N. gonorrhoeae is endemic. Quantifying the relationship between N. gonorrhoeae circulating in England and internationally also could help parameterize mathematical models exploring the relative contribution of importation and de novo development on AMR prevalence and distribution.
Rapid molecular tests for genetic markers that are highly predictive of an antimicrobial susceptibility phenotype could lead to more effective use of antimicrobials. Tests detecting markers of ciprofloxacin and cephalosporin antimicrobial susceptibility are already in development (8,29,30). However, as found in our study and elsewhere, the association between genotype and phenotype is much stronger for ciprofloxacin resistance than for cephalosporin resistance (4,31,32). Most rapid tests for cephalosporin resistance focus on detecting mutations in the penA allele, but reduced susceptibility to cephalosporins also can be caused by mutations in the semimosaic penA allele, penA-35, the nonmosaic penA allele, penA-44, or even in the mtrR and penB genes (30,33).
Molecular tests that focus on the presence or absence of 1 mutation without considering the additive effect of multiple mutations could be insufficient for detecting resistance and predicting treatment failure. Epistasis, in which phenotypic resistance is dependent on complex interactions of multiple mutant genes, is known to occur in N. gonorrhoeae (22,31,34). In our study, the antimicrobial susceptibility of isolates with identical genetic markers of resistance varied by >2 doubling dilutions, and most isolates with resistance markers were sensitive. Nonetheless, the presence of 1 mutation that belongs to a complex of mutations required for resistance indicates the potential for phenotypic resistance to develop. Clinicians could prioritize patients infected with these strains for a test of cure or consider use of alternative antimicrobial drugs unaffected by the resistance marker. In any event, mathematical modeling studies have shown that molecular tests should only be implemented if they are highly sensitive; otherwise, they could accelerate the spread of AMR (23). Elucidation of the mechanisms and genomic markers of cephalosporin resistance is needed and can be achieved through a combination of microbiologic and genomic studies, including genome wide association studies. WGS cannot replace phenotypic testing for all antimicrobial susceptibility because it can only detect known mutations associated with resistance, and novel mutations associated with resistance develop constantly in N. gonorrhoeae (31).
In conclusion, phylogenetic analyses with WGS data revealed transmission patterns of N. gonorrhoeae with reduced susceptibility in England that would not have been identified by using only epidemiologic and phenotypic data. Reduced susceptibility to antimicrobial drugs likely has emerged and spread independently in different sexual networks in England through multiple de novo mutation and recombination events and through some repeated importation by persons who have travel-associated sexual partnerships. Consequently, public health actions to limit dissemination of AMR in England should aim to reduce risk behaviors that support N. gonorrhoeae transmission and encompass the diffuse distribution and epidemiologic diversity of the population groups affected.
Dr. Town was the senior surveillance scientist on the Gonococcal Resistance to Antimicrobials Surveillance Programme (GRASP) at Public Health England and graduated from University College London with a PhD in epidemiology. Dr. Town’s research focuses on investigating new methods for understanding, monitoring, and preventing the spread of Neisseria gonorrhoeae and antimicrobial resistance.
Acknowledgments
We thank our funders, the Blood-Borne and Sexually Transmitted Infections Steering Committee of the National Institute for Health Research Health Protection Research Unit (NIHR HPRU), Caroline Sabin, Anthony Nardone, Catherine Mercer, Gwenda Hughes, Jackie Cassell, Greta Rait, Samreen Ijaz, Tim Rhodes, Kholoud Porter, Sema Mandal, and William Rosenberg; the Wellcome Trust (grant no. 098051); and Public Health England. We also thank the clinical and laboratory staff who contributed to GRASP and Hester Allen for her help with data management and analysis.
This research was funded by the National Institute for Health Research (NIHR) Health Protection Research Unit in Blood Borne and Sexually Transmitted Infections at University College London in partnership with Public Health England, in collaboration with the London School of Hygiene & Tropical Medicine. The views expressed are those of the authors and not necessarily those of the NIHR, the Department of Health and Social Care, or Public Health England.
References
- Tapsall JW. Antimicrobial resistance in Neisseria gonorrhoeae. Geneva: World Health Organization; 2001.
- Wi T, Lahra MM, Ndowa F, Bala M, Dillon JR, Ramon-Pardo P, et al. Antimicrobial resistance in Neisseria gonorrhoeae: Global surveillance and a call for international collaborative action. PLoS Med. 2017;14:
e1002344 . DOIPubMedGoogle Scholar - Demczuk W, Lynch T, Martin I, Van Domselaar G, Graham M, Bharat A, et al. Whole-genome phylogenomic heterogeneity of Neisseria gonorrhoeae isolates with decreased cephalosporin susceptibility collected in Canada between 1989 and 2013. J Clin Microbiol. 2015;53:191–200. DOIPubMedGoogle Scholar
- Grad YH, Harris SR, Kirkcaldy RD, Green AG, Marks DS, Bentley SD, et al. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000–2013. J Infect Dis. 2016;214:1579–87. DOIPubMedGoogle Scholar
- Harris SR, Cole MJ, Spiteri G, Sánchez-Busó L, Golparian D, Jacobsson S, et al.; Euro-GASP study group. Public health surveillance of multidrug-resistant clones of Neisseria gonorrhoeae in Europe: a genomic survey. Lancet Infect Dis. 2018;18:758–68. DOIPubMedGoogle Scholar
- Town K, Obi C, Quaye N, Chisholm S, Hughes G; GRASP Collaborative Group. Drifting towards ceftriaxone treatment failure in gonorrhoea: risk factor analysis of data from the Gonococcal Resistance to Antimicrobials Surveillance Programme in England and Wales. Sex Transm Infect. 2017;93:39–45. DOIPubMedGoogle Scholar
- Kirkcaldy RD, Zaidi A, Hook EW III, Holmes KK, Soge O, del Rio C, et al. Neisseria gonorrhoeae antimicrobial resistance among men who have sex with men and men who have sex exclusively with women: the Gonococcal Isolate Surveillance Project, 2005-2010. Ann Intern Med. 2013;158:321–8. DOIPubMedGoogle Scholar
- Allan-Blitz L-T, Humphries RM, Hemarajata P, Bhatti A, Pandori MW, Siedner MJ, et al. Implementation of a rapid genotypic assay to promote targeted ciprofloxacin therapy of Neisseria gonorrhoeae in a large health system. Clin Infect Dis. 2017;64:1268–70.PubMedGoogle Scholar
- Grad YH, Kirkcaldy RD, Trees D, Dordel J, Harris SR, Goldstein E, et al. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect Dis. 2014;14:220–6. DOIPubMedGoogle Scholar
- Town K, Bolt H, Croxford S, Cole M, Harris S, Field N, et al. Neisseria gonorrhoeae molecular typing for understanding sexual networks and antimicrobial resistance transmission: A systematic review. J Infect. 2018;76:507–14. DOIPubMedGoogle Scholar
- Fifer H, Cole M, Hughes G, Padfield S, Smolarchuk C, Woodford N, et al. Sustained transmission of high-level azithromycin-resistant Neisseria gonorrhoeae in England: an observational study. Lancet Infect Dis. 2018;18:573–81. DOIPubMedGoogle Scholar
- Eyre DW, Town K, Street T, Barker L, Sanderson N, Cole MJ, et al. Detection in the United Kingdom of the Neisseria gonorrhoeae FC428 clone, with ceftriaxone resistance and intermediate resistance to azithromycin, October to December 2018. Euro Surveil. 2019;24.
- De Silva D, Peters J, Cole K, Cole MJ, Cresswell F, Dean G, et al. Whole-genome sequencing to determine transmission of Neisseria gonorrhoeae: an observational study. Lancet Infect Dis. 2016;16:1295–303. DOIPubMedGoogle Scholar
- Hughes G, Nichols T, Ison CA. Estimating the prevalence of gonococcal resistance to antimicrobials in England and Wales. Sex Transm Infect. 2011;87:526–31. DOIPubMedGoogle Scholar
- Public Health England. Gonococcal Resistance to Antimicrobials Surveillance Programme (GRASP): protocol. 2014 [22 Jan 2020]. https://www.gov.uk/government/publications/gonococcal-resistance-to-antimicrobials-surveillance-programme-grasp-protocol/gonococcal-resistance-to-antimicrobials-surveillance-programme-grasp-protocol
- Mohammed H, Ison CA, Obi C, Chisholm S, Cole M, Quaye N, et al.; GRASP Collaborative Group. Frequency and correlates of culture-positive infection with Neisseria gonorrhoeae in England: a review of sentinel surveillance data. Sex Transm Infect. 2015;91:287–93. DOIPubMedGoogle Scholar
- Cole MJ, Quaye N, Jacobsson S, Day M, Fagan E, Ison C, et al. Ten years of external quality assessment (EQA) of Neisseria gonorrhoeae antimicrobial susceptibility testing in Europe elucidate high reliability of data. BMC Infect Dis. 2019;19:281. DOIPubMedGoogle Scholar
- Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:
e15 . DOIPubMedGoogle Scholar - Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50. DOIPubMedGoogle Scholar
- Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, Keane JA, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3:
e000131 . DOIPubMedGoogle Scholar - Sánchez-Busó L, Golparian D, Corander J, Grad YH, Ohnishi M, Flemming R, et al. The impact of antimicrobials on gonococcal evolution. Nat Microbiol. 2019;4:1941–50. DOIPubMedGoogle Scholar
- Unemo M, Nicholas RA. Emergence of multidrug-resistant, extensively drug-resistant and untreatable gonorrhea. Future Microbiol. 2012;7:1401–22. DOIPubMedGoogle Scholar
- Fingerhuth SM, Bonhoeffer S, Low N, Althaus CL. Antibiotic-resistant Neisseria gonorrhoeae spread faster with more treatment, not more sexual partners. PLoS Pathog. 2016;12:
e1005611 . DOIPubMedGoogle Scholar - Xiridou M, Soetens LC, Koedijk FD, VAN DER Sande MA, Wallinga J. Public health measures to control the spread of antimicrobial resistance in Neisseria gonorrhoeae in men who have sex with men. Epidemiol Infect. 2015;143:1575–84. DOIPubMedGoogle Scholar
- Abrams AJ, Kirkcaldy RD, Pettus K, Fox JL, Kubin G, Trees DL. A Case of decreased susceptibility to ceftriaxone in Neisseria gonorrhoeae in the absence of a mosaic penicillin-binding protein 2 (penA) allele. Sex Transm Dis. 2017;44:492–4. DOIPubMedGoogle Scholar
- Al Suwayyid BA, Coombs GW, Speers DJ, Pearson J, Wise MJ, Kahler CM. Genomic epidemiology and population structure of Neisseria gonorrhoeae from remote highly endemic Western Australian populations. BMC Genomics. 2018;19:165. DOIPubMedGoogle Scholar
- Hui BB, Whiley DM, Donovan B, Law MG, Regan DG; GRAND Study Investigators. Identifying factors that lead to the persistence of imported gonorrhoeae strains: a modelling study. Sex Transm Infect. 2017;93:221–5. DOIPubMedGoogle Scholar
- Goldstein E, Kirkcaldy RD, Reshef D, Berman S, Weinstock H, Sabeti P, et al. Factors related to increasing prevalence of resistance to ciprofloxacin and other antimicrobial drugs in Neisseria gonorrhoeae, United States. Emerg Infect Dis. 2012;18:1290–7. DOIPubMedGoogle Scholar
- Donà V, Smid JH, Kasraian S, Egli-Gany D, Dost F, Imeri F, et al. Mismatch amplification mutation assay-based real-time PCR for rapid detection of Neisseria gonorrhoeae and antimicrobial resistance determinants in clinical specimens. J Clin Microbiol. 2018;56:e00365–18. DOIPubMedGoogle Scholar
- Zhao L, Liu A, Li R, Zhao S. Multiplex TaqMan real-time PCR platform for detection of Neisseria gonorrhoeae with decreased susceptibility to ceftriaxone. Diagn Microbiol Infect Dis. 2019;93:299–304. DOIPubMedGoogle Scholar
- Unemo M, Shafer WM. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev. 2014;27:587–613. DOIPubMedGoogle Scholar
- Chen SC, Yin YP, Dai XQ, Unemo M, Chen XS. First nationwide study regarding ceftriaxone resistance and molecular epidemiology of Neisseria gonorrhoeae in China. J Antimicrob Chemother. 2016;71:92–9. DOIPubMedGoogle Scholar
- Zhao S, Duncan M, Tomberg J, Davies C, Unemo M, Nicholas RA. Genetics of chromosomally mediated intermediate resistance to ceftriaxone and cefixime in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 2009;53:3744–51. DOIPubMedGoogle Scholar
- Tomberg J, Unemo M, Davies C, Nicholas RA. Molecular and structural analysis of mosaic variants of penicillin-binding protein 2 conferring decreased susceptibility to expanded-spectrum cephalosporins in Neisseria gonorrhoeae: role of epistatic mutations. Biochemistry. 2010;49:8062–70. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: February 04, 2020
Table of Contents – Volume 26, Number 3—March 2020
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Katy Town, Public Health England, 61 Colindale Ave, London NW9 5EQ, UK
Top