Volume 26, Number 8—August 2020
Research
Population Genomic Structure and Recent Evolution of Plasmodium knowlesi, Peninsular Malaysia
Figure 1
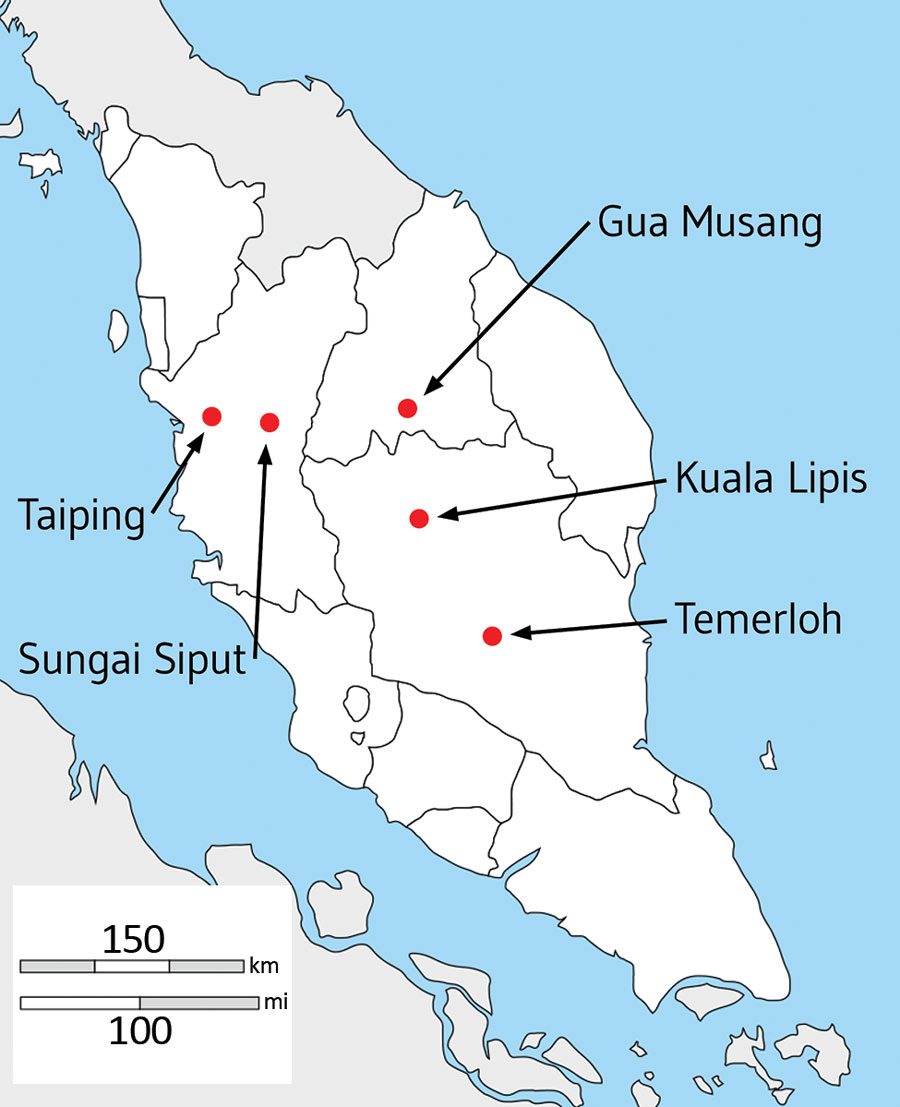
Figure 1. Locations of hospitals in peninsular Malaysia from which clinical Plasmodium knowlesi infections were sampled and sequenced in the states of Perak (Taiping and Sungai Siput), Kelantan (Gua Musang), and Pahang (Kuala Lipis and Temerloh). Of 56 infection samples processed through leukocyte depletion and subsequent DNA extraction, 32 had sufficient quantity and purity of P. knowlesi DNA for Illumina sequencing (https://www.illumina.com), of which 28 yielded high coverage genomewide sequence for population genomic analysis (sample and sequencing details listed in Appendix 1 Table).
Page created: May 18, 2020
Page updated: June 25, 2020
Page reviewed: June 25, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.