Volume 26, Number 8—August 2020
Research
Population Genomic Structure and Recent Evolution of Plasmodium knowlesi, Peninsular Malaysia
Figure 6
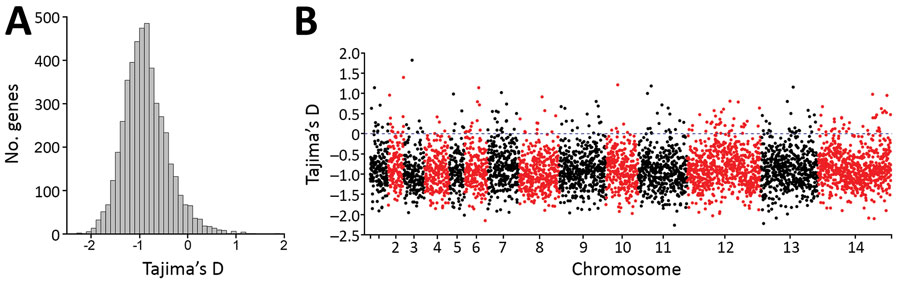
Figure 6. Summary of nucleotide site allele frequency distributions by Tajima’s D indices for all 4,742 Plasmodium knowlesi genes with >3 SNPs among the 28 cluster 3 P. knowlesi infections in Peninsular Malaysia. A) Overall values were negatively skewed with a mean Tajima’s D of −0.86, consistent with a pattern that would be caused by long-term population size expansion. B) Data for all individual genes show that those with high Tajima’s D values are distributed throughout the genome. Some of these genes are likely to be underbalancing selection (individual values for all genes are shown in Appendix 2 Datasheet 2).
Page created: May 18, 2020
Page updated: June 25, 2020
Page reviewed: June 25, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.