Volume 27, Number 3—March 2021
Research Letter
Genomic and Pathologic Findings for Prototheca cutis Infection in Cat
Cite This Article
Citation for Media
Abstract
Severe nasal Prototheca cutis infection was diagnosed postmortem for an immunocompetent cat with respiratory signs. Pathologic examination and whole-genome sequencing identified this species of algae, and susceptibility testing determined antimicrobial resistance patterns. P. cutis infection should be a differential diagnosis for soft tissue infections of mammals.
Prototheca spp. (phylum Chlorophyta, order Chlorellales, family Chlorellaceae) are ubiquitous algal organisms that represent emerging infectious agents of humans and animals (1). Protothecosis has been increasingly reported for immunocompromised human and animal patients (1,2). At least 14 species of Prototheca have been recognized; 1 case of P. cutis–associated dermatitis in an immunocompromised man has been reported (3,4). We describe a case of P. cutis in a domestic cat in Georgia, USA.
In January 2020, an 11-year-old, 5.8-kg, neutered male, domestic cat was examined for sneezing, wheezing, congestion, and rhinitis. This indoor/outdoor cat was negative for feline leukemia and feline immunodeficiency viruses. The cat showed no response to treatment with steroids and cefovecin sodium (Convenia; Zoetis, https://www.zoetis.com). From June 2019 through January 2020, the nasal planum became rounded and disfigured. A biopsy sample submitted to a private diagnostic laboratory indicated a fungal infection containing organisms suggestive of Cryptococcus spp. Because of concerns over the zoonotic potential of Cryptococcus spp., the cat was euthanized and submitted for postmortem examination.
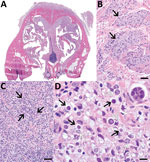
Gross reflection of the skin revealed that a locally extensive area of connective tissue and musculature overlying ≈70% of the nasal bridge and dorsal nasal planum was diffusely soft, variably tan to light orange, and mildly gelatinous (Appendix Figure 1). Microscopic evaluation revealed a severe granulomatous nodular dermatitis, panniculitis, cellulitis, pyogranulomatous osteomyelitis, and rhinitis (Figure). Disseminated throughout the nasal turbinates were numerous free and intrahistiocytic sporangia with and without endospores (Figure). Although these findings suggest previously resolved infection in the skin, subcutis, and muscle and active infection in the nasal turbinates, the initial site of infection (cutaneous vs. nasal turbinates) and disease pathogenesis could not be definitively determined (Appendix).
Fungal culture yielded white colonies growing in the presence and absence of light at 30°C (Appendix Figure 2). Cytologic examination revealed colonies of round cells with internal septations and thick walls resembling sporangia and endospores, which were identified with lactophenol cotton blue stain (Appendix Figure 2). Sporangia were gram positive, although they appeared to be unevenly stained (Appendix Figure 2). Genus and species were not identified by matrix-assisted laser desorption ionization/time-of-flight mass spectrometry and GEN III Microbial identification (Biolog, https://www.biolog.com). Partial sequencing of the internal transcribed spacer region and the D1/D2 region of the 28S rRNA gene yielded sequences 96% and 99% homologous to those from P. cutis available in BLAST (https://blast.ncbi.nlm.nih.gov) and CBS-KNAW (https://www.knaw.nl, currently Westerdijk Fungal Biodiversity Institute) databases.
Because the mitochondrial cytb gene potentially represents a new standard method for identifying Prototheca species (5), we performed whole-genome sequencing to investigate cytb as well as other genes by using Illumina MiSeq (https://www.illumina.com) (Appendix). The nuclear genome was 19,237,076-bp long, and the plastid genome was 51,673-bp long, which corresponds to genome sizes obtained from sequencing of P. cutis JCM15793 strain ATCC PRA-138 (https://www.atcc.org) (6). We submitted the genome assembly to GenBank (accession no. JABBYS000000000). The in silico–targeted gene alignment revealed genes homologous with available P. cutis sequences including cytB (99.8%, accession no. MT363977), chloroplast genome DNA (99.54%), 18S rRNA gene (100%, accession no. MT360051), ITS (99.12%, accession no. MT359908), and 28S rRNA (D1/D2 domain) (100%, accession no. MT360265), which were deposited in GenBank.
We performed susceptibility testing for antifungal and antimicrobial drugs because both have been used against Prototheca spp. infections in animals (7,8). The isolate showed high MICs for fluconazole and itraconazole and a low MIC for amphotericin B (Table). Resistance to fluconazole and susceptibility to amphotericin B are well recognized for other Prototheca species (9); however, resistance to itraconazole could be unique to P. cutis. MICs were high for most antimicrobial drugs, including cefovecin, which had been unsuccessful in treating the cat (Table). Oxacillin, pradofloxacin, and trimethoprim/sulfamethoxazole inhibited growth at the lowest concentrations, indicating in vitro sensitivity to these drugs. We further investigated whether resistance genes from the whole-genome sequence corresponded to phenotypic resistance. We used the Comprehensive Antibiotic Resistance Database (https://card.mcmaster.ca) to identify genes conferring resistance to β-lactams, tetracyclines, aminoglycosides, chloramphenicol, and vancomycin. Consistent with MICs, no resistance genes corresponded to pradofloxacin and trimethoprim/sulfamethoxazole (Table). The MIC data provided here may be helpful for establishing future clinical breakpoints for Prototheca spp.
Our primary concern with regard to this case was determining the zoonotic potential of the agent, which was initially misdiagnosed as Cryptococcus spp. Although Prototheca spp. are widely considered to be zoonotic agents, reports of definitive cases of zoonotic transmission are lacking in the literature. Zoonotic transmission from bovids is thought to occur via consumption of contaminated milk (10). The zoonotic potential of P. cutis is unclear; infectivity is probably similar to that of other Prototheca spp.
Our report of P. cutis isolation from a cat reinforces protothecosis as an emerging infectious disease of humans and animals. We emphasize the potential of P. cutis to infect presumably immunocompetent hosts. The veterinary and human medical communities should be aware of the unusual clinical, pathologic, and microbiological manifestations of protothecosis.
Dr. Maboni is a board-certified veterinary microbiologist working as an assistant professor at the University of Guelph, Canada. Her primary research interests are medical diagnosis of bacterial and fungal diseases.
Acknowledgment
We thank the talented histology and microbiology technicians at the Athens Veterinary Diagnostic Laboratory for their technical assistance, especially Paula Bartlett and Amy McKinney.
References
- Pal M, Abraha A, Rahman MT, Dave P. Protothecosis: an emerging algal disease of humans and animals [cited 2020 Jun 17]. https://www.researchgate.net/publication/266359627_Protothecosis_an_emerging_algal_disease_of_humans_and_animals
- Lanotte P, Baty G, Senecal D, Dartigeas C, Bailly E, Duong TH, et al. Fatal algaemia in patient with chronic lymphocytic leukemia. Emerg Infect Dis. 2009;15:1129–30. DOIPubMedGoogle Scholar
- Jagielski T, Bakula Z, Gawor J, Maciszewski K, Kusber WH, Dylag M, et al. The genus Prototheca (Trebouxiophyceae, Chlorophyta) revisited: implications from molecular taxonomic studies. Algal Res. 2019;43:
101639 . DOIGoogle Scholar - Satoh K, Ooe K, Nagayama H, Makimura K. Prototheca cutis sp. nov., a newly discovered pathogen of protothecosis isolated from inflamed human skin. Int J Syst Evol Microbiol. 2010;60:1236–40. DOIPubMedGoogle Scholar
- Jagielski T, Gawor J, Bakuła Z, Decewicz P, Maciszewski K, Karnkowska A. cytb as a new genetic marker for differentiation of Prototheca species. J Clin Microbiol. 2018;56:e00584–18. DOIPubMedGoogle Scholar
- Suzuki S, Endoh R, Manabe RI, Ohkuma M, Hirakawa Y. Multiple losses of photosynthesis and convergent reductive genome evolution in the colourless green algae Prototheca. Sci Rep. 2018;8:940. DOIPubMedGoogle Scholar
- Pressler BM, Gookin JL, Sykes JE, Wolf AM, Vaden SL. Urinary tract manifestations of protothecosis in dogs. J Vet Intern Med. 2005;19:115–9. DOIPubMedGoogle Scholar
- Vince AR, Pinard C, Ogilvie AT, Tan EO, Abrams-Ogg AC. Protothecosis in a dog. Can Vet J. 2014;55:950–4.PubMedGoogle Scholar
- Marques S, Silva E, Carvalheira J, Thompson G. Short communication: In vitro antimicrobial susceptibility of Prototheca wickerhamii and Prototheca zopfii isolated from bovine mastitis. J Dairy Sci. 2006;89:4202–4. DOIPubMedGoogle Scholar
- Melville PA, Watanabe ET, Benites NR, Ribeiro AR, Silva JA, Garino Junior F, et al. Evaluation of the susceptibility of Prototheca zopfii to milk pasteurization. Mycopathologia. 1999;146:79–82. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: February 14, 2021
1These authors contributed equally to this article.
Table of Contents – Volume 27, Number 3—March 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Grazieli Maboni, Department of Pathobiology, Ontario Veterinary College, University of Guelph, 50 Stone Rd E, Guelph, ON N1G 2W1, Canada
Top