Volume 28, Number 7—July 2022
Dispatch
Use of Human Intestinal Enteroids to Evaluate Persistence of Infectious Human Norovirus in Seawater
Cite This Article
Citation for Media
Abstract
Little data on the persistence of human norovirus infectivity are available to predict its transmissibility. Using human intestinal enteroids, we demonstrate that 2 human norovirus strains can remain infectious for several weeks in seawater. Such experiments can improve understanding of factors associated with norovirus survival in coastal waters and shellfish.
Human noroviruses are the major cause of viral gastroenteritis worldwide (1) and the most common cause of foodborne or waterborne outbreaks in Europe (2). Noroviruses spread through fecal–oral transmission, mainly person to person, but also spread through environmental contamination (1). Food and drinks can be contaminated by infected food handlers and, during production, by human sewage spillover (3). When grown in contaminated seawater, filter-feeding shellfish bioaccumulate human noroviruses in their tissues (2,4). Shellfish, especially those eaten raw, are among the main foods involved in foodborne epidemics (2,5).
Noroviruses are diverse, positive-stranded RNA viruses, classified into >10 genogroups (G) and many genotypes; most noroviruses that infect humans belong to genogroups GI and GII (6). Since 1995, the epidemiology of human noroviruses has been dominated by the GII.4 genotype (1). Of note, GII.4 appears to be predominantly transmitted person to person, whereas other genotypes, such as GII.6, GII.3, and some from GI, are more often implicated in foodborne or waterborne outbreaks (1–5). This difference might reflect variations in particle resistance to environmental conditions (1,7), but empirical data are lacking.
The small, nonenveloped human norovirus particles are considered very stable outside their host, especially in aquatic environments (1,7,8). Particles are also highly infectious, leading to human infection even when very low amounts of virus are present in shellfish (9). Yet, for almost 50 years, the lack of a reproducible cell culture system impaired the direct assessment of human norovirus infectivity in environmental conditions. Hence, data used for risk assessment rely on molecular assays or surrogate viruses (2). Previously, we used a surrogate calicivirus, Tulane virus (TuV), to estimate the persistence of infectious human norovirus in shellfish (10). However, because surrogates might underestimate the actual stability of human norovirus (11) and do not enable comparisons between different norovirus strains, direct assessments of infectivity in the environment and foods are needed to learn more about foodborne transmission and design optimal sanitary regulations (2).
Since 2016, human intestinal enteroids (HIEs) have enabled the in vitro cultivation of many human norovirus strains and represent a physiologically relevant model to assess whether the virus is infectious (12–15). In this study, we used HIEs to evaluate the persistence of infectious human norovirus in natural seawater, the last matrix before bioaccumulation by shellfish, in comparison with TuV.
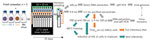
We compared the stability in seawater of 2 human norovirus strains, GII.4 (TCH11-64) and GII.3 (TCH04-577), obtained from human stool filtrates as described previously (12), and 1 TuV strain (M33) produced in simian LLC-MK2 cells (10). Ethics approval for collection of virus-containing fecal samples and human intestinal cells was obtained from the Baylor College of Medicine Institutional Review Board. We conducted 3 experiments with fresh samples of natural seawater (Table 1). We used viral stocks to spike 120 mL of seawater, which we then split into 10 mL-aliquots and incubated at 12°C in a thermostatic cabinet (Memmert, https://www.memmert.com) (Figure 1). Once or twice a week, we randomly sampled an aliquot, extracted nucleic acids from 100 µL by using the NucliSens kit on a MiniMag (bioMérieux, https://www.biomerieux.com), and assessed the viral genome concentration by quantitative reverse transcription PCR (10).
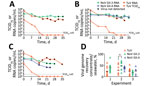
During experiments 1 and 2, the genomic concentration of human norovirus GII.3 remained highly stable; 0.8 log10 losses in experiment 1 and 1.2 log10 losses in experiment 2 occurred over 5 weeks (Figure 2, panels A, B). We did not assess GII.4 virus in experiment 1, but we observed similar stability at the genomic level in experiment 2, a 0.5 log10 decrease (Figure 2, panel B). During experiment 3, GII.3 and GII.4 genomic concentrations were >1 log10 lower than the other experiments at day 0 and reached a total loss of 1.8 and 2.7 log10 over 4 weeks (Figure 2, panel C). For the 3 experiments, TuV genomic levels were higher than human norovirus at day 0 but decreased more quickly; total losses of 2.7 (experiment 1), 3.2 (experiment 2) and 3.4 (experiment 3) log10 occurred over 5 weeks (Figure 2, panels A–C), consistent with the decay rate observed previously in contaminated oysters (10).
For the remainder (9.9 mL) of the seawater aliquots, we filter-sterilized, concentrated by centrifugal ultrafiltration, and desalted, adapting a method used to purify infectious TuV from oysters (10). To verify efficacy, we used 100 µL of purified concentrate for RNA extraction to quantify the viral genome and to calculate the proportion of virus recovered in the concentrate compared with the proportion of virus in seawater (Figure 1). For all experiments combined, viral recovery ranged from 4% to 61% for TuV, 6% to 70% for GII.3, and 4% to 37% for GII.4 (Figure 2, panel D). The recovery of human norovirus tended to be even higher than for TuV, especially in the case of GII.3 in experiment 2 (Figure 2, panel D).
We used purified concentrates of TuV to assess its infectious titer through 50% tissue culture infectious dose on LLC-MK2 cells (10) (Figure 1). Infectious TuV was detected for 14 days during experiment 1 and for 21 days during experiment 2 (Figure 2, panels A, B), similar to the length of detection in contaminated oysters (10). Experiment 3 also showed a faster loss of infectious TuV, which was not detected after 7 days (Figure 2, panel C).
We used the purified concentrates of human norovirus to infect differentiated jejunal J2 HIE monolayers in triplicate (12), either upon collection or after storage at −80°C (Figure 1). The geometric mean fold increase (GMFI) in viral genome was measured between 1 hour and 72 hours after infection; the virus was considered infectious when GMFI >3.0 (Table 2). We detected infectious human norovirus GII.3 at up to 28 days (experiment 1), 31 days (experiment 2), and 14 days (experiment 3); infectious norovirus GII.4 was recovered throughout the 35 days in experiment 2 and through day 7 in experiment 3 (Table 2). Progressive loss in human norovirus infectivity is suggested by the GMFI decrease during all experiments for both viruses (Table 2). Of note, for all experiments, infectious GII.3 and GII.4 were detected for longer periods of time than infectious TuV (Figure 2, panels A–C), suggesting that human norovirus is more stable than TuV in seawater, especially because the initial concentrations of TuV were higher (Figure 2, panels A–C). Our results also suggest that the persistence of GII.3 and GII.4 is similar in these settings, but this finding needs further validation with a quantitative assay, because J2 HIE monolayers are more susceptible to GII.4 than GII.3 (12). Indeed, we observed the absence of infectious human norovirus when input genome levels were close to the sensitivity threshold of the assay (2 × 104 for GII.3, 1.2 × 103 for GII.4) (12), which suggests that infectious human norovirus particles might still have been present but were undetected. Finally, all virus data show that experiment 3 differs from the 2 others, which could have been caused by uncharacterized variables of the different seawater samples.
This study demonstrates that HIEs can be used to study infectious human norovirus persistence in seawater, an environmental matrix, and confirms the virus’s high stability. Using 3 natural seawater samples, we observed persistent yet variable viability of human norovirus, showing that the nature of the seawater affects viral infectivity. This model will enable further research assessing possible factors at play, such as the bacterial flora or the physio-chemical parameters of the water. Together with data on foodborne outbreaks, this model will help determine the behavior of human norovirus in the environment and thus protect human health by enabling sanitary regulations to be adapted for actual infectious risks.
Dr. Desdouits is an environmental virology researcher at the Health, Environment and Microbiology Laboratory at Institut Franҫais de Recherche pour l’Exploitation de la Mer (Ifremer), Nantes, France. Her main interests lie in understanding the contamination of coastal waters and shellfish with human enteric pathogens such as norovirus, and how virus diversity and persistence contribute to the foodborne transmission of these pathogens.
Acknowledgments
We are thankful to Jacques Le Pendu for helpful discussions, sharing reagents, and technical help with norovirus and enteroids.
This work was funded by the Direction Générale de l’Alimentation (DGAl) and the Agence Nationale de la Recherche (project GOyAVE, n°19-CE35-0014-01).
References
- de Graaf M, van Beek J, Koopmans MPG. Human norovirus transmission and evolution in a changing world. Nat Rev Microbiol. 2016;14:421–33. DOIPubMedGoogle Scholar
- Savini F, Giacometti F, Tomasello F, Pollesel M, Piva S, Serraino A, et al. Assessment of the impact on human health of the presence of norovirus in bivalve molluscs: what data do we miss? Foods. 2021;10:2444. DOIPubMedGoogle Scholar
- Mathijs E, Stals A, Baert L, Botteldoorn N, Denayer S, Mauroy A, et al. A review of known and hypothetical transmission routes for noroviruses. Food Environ Virol. 2012;4:131–52. DOIPubMedGoogle Scholar
- Le Guyader FS, Atmar RL, Le Pendu J. Transmission of viruses through shellfish: when specific ligands come into play. Curr Opin Virol. 2012;2:103–10. DOIPubMedGoogle Scholar
- Marsh Z, Shah MP, Wikswo ME, Barclay L, Kisselburgh H, Kambhampati A, et al. Epidemiology of foodborne norovirus outbreaks—United States, 2009–2015. Food Saf (Tokyo). 2018;6:58–66. DOIPubMedGoogle Scholar
- Chhabra P, de Graaf M, Parra GI, Chan MC-W, Green K, Martella V, et al. Updated classification of norovirus genogroups and genotypes. J Gen Virol. 2019;100:1393–406. DOIPubMedGoogle Scholar
- Pogan R, Dülfer J, Uetrecht C. Norovirus assembly and stability. Curr Opin Virol. 2018;31:59–65. DOIPubMedGoogle Scholar
- Boehm AB, Silverman AI, Schriewer A, Goodwin K. Systematic review and meta-analysis of decay rates of waterborne mammalian viruses and coliphages in surface waters. Water Res. 2019;164:
114898 . DOIPubMedGoogle Scholar - Teunis PFM, Le Guyader FS, Liu P, Ollivier J, Moe CL. Noroviruses are highly infectious but there is strong variation in host susceptibility and virus pathogenicity. Epidemics. 2020;32:
100401 . DOIPubMedGoogle Scholar - Polo D, Schaeffer J, Teunis P, Buchet V, Le Guyader FS. Infectivity and RNA persistence of a norovirus surrogate, the Tulane virus, in oysters. Front Microbiol. 2018;9:716. DOIPubMedGoogle Scholar
- Manuel CS, Moore MD, Jaykus LA. Predicting human norovirus infectivity - Recent advances and continued challenges. Food Microbiol. 2018;76:337–45. DOIPubMedGoogle Scholar
- Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–93. DOIPubMedGoogle Scholar
- Costantini V, Morantz EK, Browne H, Ettayebi K, Zeng XL, Atmar RL, et al. Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerg Infect Dis. 2018;24:1453–64. DOIPubMedGoogle Scholar
- Chan MC, Cheung SKC, Mohammad KN, Chan JCM, Estes MK, Chan PKS. Use of human intestinal enteroids to detect human norovirus infectivity. Emerg Infect Dis. 2019;25:1730–5. DOIPubMedGoogle Scholar
- Overbey KN, Zachos NC, Coulter C, Jacangelo J, Schwab KJ. Recovery of infectious human norovirus GII.4 Sydney from fomites via replication in human intestinal enteroids. Front Cell Infect Microbiol. 2021;11:
693090 . DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: June 09, 2022
1These first authors contributed equally to this article.
2Current affiliation: Centro de Investigaciones Biologicas–Facultade de Bioloxía & CRETUS, Universidade de Santiago de Compostela, Santiago de Compostela, Spain.
3Current affiliation: SECALIM UMR 104 Oniris/Inrae, Nantes, France.
Table of Contents – Volume 28, Number 7—July 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Marion Desdouits, Ifremer, rue de l’Ile d’Yeu, 44311 Nantes CEDEX 03, France
Top