Volume 29, Number 7—July 2023
Research
Effect of Norovirus Inoculum Dose on Virus Kinetics, Shedding, and Symptoms
Abstract
The effect of norovirus dose on outcomes such as virus shedding and symptoms after initial infection is not well understood. We performed a secondary analysis of a human challenge study by using Bayesian mixed-effects models. As the dose increased from 4.8 to 4,800 reverse transcription PCR units, the total amount of shed virus in feces increased from 4.5 × 1011 to 3.4 × 1012 genomic equivalent copies; in vomit, virus increased from 6.4 × 105 to 3.0 × 107 genomic equivalent copies. Onset time of viral shedding in feces decreased from 1.4 to 0.8 days, and time of peak viral shedding decreased from 2.3 to 1.5 days. Time to symptom onset decreased from 1.5 to 0.8 days. One type of symptom score increased. An increase in norovirus dose was associated with more rapid shedding and symptom onset and possibly increased severity. However, the effect on virus load and shedding was inconclusive.
Norovirus is a major cause of foodborne disease and causes a large number of cases, hospitalizations, and deaths in the United States and globally (1–4). Specific treatments are not available, and vaccines are still under development (4,5). Generic infection control measures are the best approaches to minimizing disease burden (6–10).
An increase in exposure dose (number of virus particles) is associated with an increased risk for infection; this principle applies to norovirus (11–14) and many other pathogens (15,16). Less is known about the possible effect of dose on infection outcomes after infection has occurred. For acute infections such as influenza, infectious bronchitis virus, and parainfluenza virus, animal studies and models suggest that dose influences the virus load kinetics (17–19). For norovirus, some evidence from experimental challenge studies suggests that dose is associated with more rapid onset of symptoms (20). To further elucidate the effect of inoculum dose on infection outcomes such as virus shedding and symptom severity, we performed a secondary analysis of data from a human norovirus challenge study (20).
In this article, we will give brief descriptions of our methods. We have also provided complete modeling and analysis details, including all data and code needed to reproduce our results (Appendix).
Data
The data we used for our analyses are from a human challenge study registered at ClinicalTrials.gov (trial no. NCT00138476) (20–24). The clinical protocol was reviewed and approved by the institutional review boards of the Baylor College of Medicine and The Houston Methodist Hospital, and written informed consent was obtained from each study participant.
In the challenge study, 57 healthy persons (18–50 years of age) were randomly inoculated with either placebo or norovirus genogroup I genotype 1 strain (GI.1 NV) at 4 different doses (0.48, 4.8, 48, or 4,800 reverse transcription PCR [RT-PCR] units). Of the 21 persons who became infected, 1 person was unavailable for follow-up, and thus we excluded that patient from all analyses. In addition, only 1 person in the 0.48-unit dose group became infected, so we excluded this person from our main analyses. Therefore, remaining for our analysis were 6 persons in the 4.8-unit dose group, 7 persons in the 4.8-unit dose group, and 6 persons in the 4,800-unit dose group. We provide analyses that include the 1 person who was infected at the 0.48-dose level (Appendix).
All persons were isolated in the research center for >4 days (96 hours) after inoculation. The study personnel collected samples of feces and vomit and recorded clinical symptoms.; samples were also collected for 4–8 weeks postinoculation. For some of our analyses, we focused on the 96 hours during which persons were under clinical observation. For other analyses, we included the data collected after persons returned home. We state which data are used for each analysis.
Overall Analysis Approach and Implementation
Because we performed a secondary data analysis, a strict null hypothesis significance testing framework using p values was not suitable, so we performed all analyses in a Bayesian framework. For all analyses, we used Bayesian mixed-effects models. We treated the dose as a continuous variable for the results shown in the article. We also provide a sensitivity analysis with dose modeled as categorical (Appendix). We report the mean of the estimated posterior distribution with 95% equal-tailed credible intervals (CrIs) for all model results (25). We conducted all analyses using R version 4.2.3 (26), and Stan (27), accessed through the brms package in R (28). We used Rhat values to diagnose convergence (28).
Analysis of Virus Shedding Outcomes
We measured virus shedding concentration in samples by either an immunomagnetic capture (IMC) RT-PCR, which provided a qualitative readout (positive or negative), or real-time quantitative RT-PCR (qRT-PCR), which provided a quantitative readout in genomic equivalent copies (GEC) (21). Those 2 methods had limits of detection (LOD) at 15,000 GEC/g of stool (LOD1) and 40,000,000 GEC/g of stool (LOD2). Therefore, the virus shedding concentration could be between 0 and LOD1 (negative IMC, negative qRT-PCR), between LOD1 and LOD2 (positive IMC, negative qRT-PCR), or a quantitative measurement above LOD2 (positive qRT-PCR). We reported vomit shedding data similarly, with either a numeric value or a positive or negative readout. We accounted for this censored data structure in our models (Appendix).
We obtained the total virus contained in each sample by multiplying virus concentration by sample weight for feces (GEC/g × weight of feces in grams) or sample volume for vomit (GEC/mL × volume of vomit in mL). We calculated each participant’s total amount of virus shedding in feces and vomit by summing virus shedding values for all samples per participant. We used a linear model structure to analyze associations between inoculum dose and the total amount of virus shedding.
In a further analysis, we modeled the longitudinal time-series of virus concentration in feces, V(t), using the 4-parameter equation
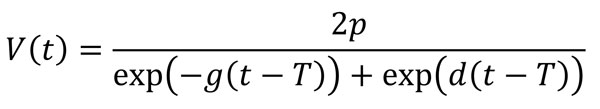
which was shown to accurately describe trajectories of acute viral infections (17,29). We fitted the trajectories by using a Bayesian nonlinear mixed-effects model in which the mean of the response was described using this equation. We used the comparison between the parameter’s prior and posterior distributions to ensure that the choice of prior distribution had no significant effect on our results. We sampled from the posterior distribution of the estimated parameters to obtain predicted trajectories of virus concentration kinetics. From those time-series, we computed several summary quantities: virus shedding onset (time at which the trajectory crossed LOD1); time to peak virus shedding; shedding duration, defined as the total amount of time at which virus concentration was above LOD1; and total amount of virus shed, defined as the area under the virus concentration curve.
Vomiting episodes were too few (11 persons with 26 samples of vomit) to enable a time-series analysis similar to the one we performed for virus shedding in feces. We have compiled vomit event time-series data (Appendix).
Analysis of Symptom Outcomes
The study recorded 10 kinds of symptoms: body temperature, malaise, muscle aches, headache, nausea, chills, anorexia, cramps, unformed or liquid feces, and vomiting. Clinical symptom scores (except feces and vomit) were reported as none = 0, mild = 1, moderate = 2, or severe = 3. For feces, we used a scoring of solid = 0, unformed = 1, and liquid = 2. Vomit was reported as absent = 0 or present = 1.
We considered time to symptom onset (incubation period) and 2 symptom scores as outcomes of interest. We defined time to system onset as the time from inoculation to the first reported symptom of any type. For the first symptom score, we used a modified Vesikari score (MVS) that was previously applied to measure norovirus severity (5,30–33). We computed the MVS by using a limited number of symptoms (i.e., fever, diarrhea, and vomiting). We also developed an additional score, which we call the comprehensive symptom score (CSS), which encompasses all reported 10 symptoms in this study. Additional details of score computation, scores for each individual, and further model details are provided (Appendix).
Sensitivity Analyses
We performed 2 sensitivity analyses. In the first analysis, we treated dose as categorical rather than continuous. In the second analysis, we included 1 person who became infected after exposure to a dose of 0.48 RT-PCR units.
Data Description
Detailed descriptions of the study can be found in previous publications (20–24). We summarized characteristics of the infected persons included in our analyses (Table). Distributions of age, sex, and ABO blood group status were generally similar across dose groups.
Association between Dose and Total Virus Shedding
We computed total virus shedding in either feces or vomit by summing the amount of shed virus in all samples for each person. We focused on fecal shedding during the first 96 hours of the study, when patients were under clinical observation. Almost all viral shedding events that occurred during this timeframe were recorded. Every person shed virus in >1 fecal sample. All vomiting events occurred within the first 96 hours, and only 11 persons vomited. Virus shedding showed some association with dose, although with a fair amount of uncertainty (Figure 1), leading to overall inconclusive results. We developed an alternative analysis using fecal shedding that includes the self-reported data after persons returned to their homes (Appendix). In that case, we observed no noticeable association.
Association between Dose and Viral Kinetics
Next, we fitted the virus concentration model to the time-series data for virus load for each person. The parameter’s prior and posterior distributions showed that the choice of prior distribution had no significant effect on our results (Appendix).
We calculated the population-level curves per dose group for the estimated virus load trajectories (Figure 2) and developed fitted curves for each person (Appendix). The curves show a trend toward more rapid onset and earlier virus peak as dose increases (Figure 2, panel B) but little effect on shedding duration and total viral load (Figure 2, panel A). To further quantify these results, we sampled trajectories from the posterior distributions. For each trajectory, we computed 4 quantities (indicated in Figure 2, panel A): shedding onset (i.e., time at which virus became detectable), time of peak virus shedding, duration of virus shedding, and the total amount of virus shed (computed as area under the curve). We then examined the distribution of each of these quantities.
We calculated the model-predicted relationship between dose and those 4 quantities (Figure 3). As the dose increased from 4.8 to 4,800 RT-PCR units, average onset time decreased from 1.4 (95% CrI 1.1–1.8) to 0.8 (95% CrI 0.5–1.1) days, and the time of virus peak decreased from 2.3 (95% CrI 2–2.8) to 1.5 (95% CrI 1.3–1.8) days. We observed a very slight trend toward increased duration of shedding, from 23.7 (95% CrI 17.8–30.6) to 26.4 (95% CrI 19–35.8) days. Total virus load barely changed, from 1.5 × 1010 (95% CrI 2.2 × 109–5.2 × 1010) to 1.7 × 1010 (95% CrI 1.9 × 109–6.6 × 1010) GEC × days/g.
Association between Dose and Symptoms
We investigated associations between dose and symptom related outcomes next. A higher inoculum dose was associated with a shorter incubation period (more rapid symptoms onset) (Figure 4). The incubation period decreased from 1.5 (95% CrI 0.9–2.5) to 0.8 (95% CrI 0.4–1.4) days as dose increased (Figure 4, panel A).
Our model estimated a slight increase in symptoms as measured by the MVS, from 2.9 (95% CrI 1.4–5.2) to 3.3 (95% CrI 1.4–6.5) as dose increased (Figure 4, panel B). The CSS showed a more pronounced increase, from 9.4 (95% CrI 6.1–13.6) to 18.7 (95% CrI 11.8–28.3) (Figure 4, panel C). A further analysis suggests that the different pattern seen for the MVS and CSS might be attributable to those symptoms that are part of the MVS not showing an association with dose, whereas a few symptoms (e.g., cramps, malaise, nausea) that are part of the CSS but not the MVS do show a correlation with dose (Appendix).
Sensitivity Analyses
We performed 2 main sensitivity analyses (Appendix). In the first sensitivity analysis, we treated dose as categorical (low, medium, or high) instead of continuous. For this analysis, total virus shedding in feces and vomit was highest at the intermediate dose, though with overlap of the credible intervals for all doses. Similar to results for the main analysis, an increase in dose led to earlier onset and peak of shedding. Duration of shedding and total virus load concentration also suggested the highest levels at intermediate doses, although again with overlap in uncertainty estimates. Symptom onset was earlier, and the CSS measure increased, with no noticeable effect on the MVS measure.
In the second sensitivity analysis, we included 1 person infected after receiving the lowest dose (0.48 RT-PCR units). For this dataset, we found similar patterns of increasing total virus shedding in feces and vomit as dose increased. Also consistent with those results, onset and peak of shedding occurred earlier but duration of shedding and total virus load concentration did not change noticeably. Symptom onset was earlier and stronger based on the CSS measure, with no noticeable effect on the MVS.
The categorical analysis suggested similar patterns but supported, albeit very tentatively, that intermediate dose might be associated with the highest level of shedding. However, because only a single person fell into the lowest-dose category, a categorical analysis that included that person did not seem to be useful, so we did not perform such an analysis.
In time series models, we treated values below the limits of detection as censors. In other virus shedding models, we additionally performed 2 sensitivity analyses to explore the effect of choices for the values that are below the limits of detection. The conclusions remained consistent (Appendix).
We explored the effect of norovirus inoculum dose on infection and disease outcomes, an important gap in the literature. We found that an increased dose was associated with a faster onset and peak of virus shedding in feces (Figure 3, panel A, B) but not with fecal shedding duration and total virus concentration (Figure 3, panel C, D). A trend toward increased total shedding was noted for both feces and vomit (Figure 1). Our analysis also showed a pattern of accelerated onset of symptoms. Symptom severity showed an increase with inoculum dose for the CSS measure but not the MVS measure (Figure 4), possibly because only some symptoms are affected by dose, and those symptoms are captured by CSS but not MVS (Appendix). An increase in symptoms despite no noticeable change in virus load suggests that symptoms are mostly immune-mediated. We found mild evidence that a high virus growth rate associated with increased symptoms (Appendix); thus, a more rapid initial virus growth might trigger a stronger immune response. This finding could be tested in studies that measure components of the ensuing immune response.
Findings similar to ours have been reported for other enteric pathogens. The clinical manifestation of typhoid illness appears to be independent of inoculum dose, whereas the onset of symptoms was more rapid after a higher infectious dose (34). More rapid onset of symptoms after a larger infectious dose has also been observed with cholera infections (35). This finding could suggest a general pattern of dose-dependent incubation periods for enteric diseases. We did not find evidence of presymptomatic virus shedding, which could be attributable to the fact that diarrhea and vomit were considered as symptoms in our research, which explains the similar time of virus shedding onset time and incubation period.
The association between dose and severity might partially explain the results of several recent norovirus vaccine candidates. Those vaccines have shown limited effectiveness at reducing the risk for infection but do seem to reduce disease outcomes (5,36). Perhaps protection induced by current vaccine candidates (assumed to be mainly mediated by antibodies) is not enough to provide sterilizing immunity and thus prevent infection but can reduce the effective dose that starts an infection and thereby reduce symptoms. This pattern would be consistent with our findings here.
However, it is unclear what the typical norovirus dose is for natural infections and how that dose compares with the doses chosen in the challenge study data we analyzed. This uncertainty limits any possibility to generalize results obtained from challenge studies to natural infections or the potential role of vaccine candidates at influencing the effective inoculum size that starts an infection. Thus, potential clinical or epidemiologic implications of changes in dose for natural infections will need to await further investigations to determine the potential applicability of challenge study results to such natural infection settings.
Our analysis was a secondary data analysis of a limited number of persons, which resulted in wide credible intervals and constrained further explorations of nonlinear models. The associations we found may not equal to causality. As such, our results should be considered exploratory and need to be confirmed in future studies. Further studies, ideally with larger sample sizes, are needed. Larger sample sizes might also allow for stratification on the basis of host characteristics, which could yield information regarding possible interactions between host characteristics and dose–outcome relationships.
In conclusion, if we can assume that the associations we found have an underlying causal relation (something that needs to be confirmed in future studies), our results suggest that norovirus dose might affect some infection outcomes while not influencing others. Thus, when comparing results across challenge studies or trying to combine data from multiple studies for analysis, some care must be taken if doses are different. In some instances, combining data across studies seems reasonable, such as combining data from multiple studies to focus on viral shedding. However, for symptom-related outcomes and quantities that focus on norovirus infection kinetics, dose differences might lead to differences between studies that prohibit easy comparison.
Dr. Ge is an assistant professor at the University of Southern Mississippi. His research focuses on infectious disease epidemiology, covering pathogens like influenza, norovirus, SARS-CoV-2, and tuberculosis.
Acknowledgment
Y.G., J.L., K.K., B.L., and A.H. are partially supported by National Institutes of Health grant no. R01 GM124280. Y.G. was supported by the Start-Up Grant from the University of Southern Mississippi. J.L. is partially supported by the National Institute of Food and Agriculture at the US Department of Agriculture (grant no. 2019-67017-29642). Z.B. is supported by the University of Georgia Graduate School. Y.S. is partially supported by National Institutes of Health grant no. R35 GM146612. D.G. and A.O. are partially supported by the Office of Research and Development Medical Research Service Department of Veterans Affairs, Public Health Service (grant no. DK56338), which funds the Texas Medical Center Digestive Diseases Center.
References
- Patel MM, Widdowson M-A, Glass RI, Akazawa K, Vinjé J, Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. 2008;14:1224–31. DOIPubMedGoogle Scholar
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128. DOIPubMedGoogle Scholar
- Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. Global economic burden of norovirus gastroenteritis. PLoS One. 2016;11:
e0151219 . DOIPubMedGoogle Scholar - Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis. 2011;17:7–15. DOIPubMedGoogle Scholar
- Atmar RL, Bernstein DI, Harro CD, Al-Ibrahim MS, Chen WH, Ferreira J, et al. Norovirus vaccine against experimental human Norwalk Virus illness. N Engl J Med. 2011;365:2178–87. DOIPubMedGoogle Scholar
- Johnston CP, Qiu H, Ticehurst JR, Dickson C, Rosenbaum P, Lawson P, et al. Outbreak management and implications of a nosocomial norovirus outbreak. Clin Infect Dis. 2007;45:534–40. DOIPubMedGoogle Scholar
- Isakbaeva ET, Widdowson M-A, Beard RS, Bulens SN, Mullins J, Monroe SS, et al. Norovirus transmission on cruise ship. Emerg Infect Dis. 2005;11:154–8. DOIPubMedGoogle Scholar
- Carling PC, Bruno-Murtha LA, Griffiths JK. Cruise ship environmental hygiene and the risk of norovirus infection outbreaks: an objective assessment of 56 vessels over 3 years. Clin Infect Dis. 2009;49:1312–7. DOIPubMedGoogle Scholar
- Wikswo ME, Cortes J, Hall AJ, Vaughan G, Howard C, Gregoricus N, et al. Disease transmission and passenger behaviors during a high morbidity Norovirus outbreak on a cruise ship, January 2009. Clin Infect Dis. 2011;52:1116–22. DOIPubMedGoogle Scholar
- Bitler EJ, Matthews JE, Dickey BW, Eisenberg JNS, Leon JS. Norovirus outbreaks: a systematic review of commonly implicated transmission routes and vehicles. Epidemiol Infect. 2013;141:1563–71. DOIPubMedGoogle Scholar
- Teunis PFM, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, et al. Norwalk virus: how infectious is it? J Med Virol. 2008;80:1468–76. DOIPubMedGoogle Scholar
- Van Abel N, Schoen ME, Kissel JC, Meschke JS. Comparison of risk predicted by multiple norovirus dose-response models and implications for quantitative microbial risk assessment. Risk Anal. 2017;37:245–64. DOIPubMedGoogle Scholar
- Messner MJ, Berger P, Nappier SP. Fractional poisson—a simple dose-response model for human norovirus. Risk Anal. 2014;34:1820–9. DOIPubMedGoogle Scholar
- Schmidt PJ. Norovirus dose-response: are currently available data informative enough to determine how susceptible humans are to infection from a single virus? Risk Anal. 2015;35:1364–83. DOIPubMedGoogle Scholar
- Haas CN. Estimation of risk due to low doses of microorganisms: a comparison of alternative methodologies. Am J Epidemiol. 1983;118:573–82. DOIPubMedGoogle Scholar
- Teunis PFM, Nagelkerke NJD, Haas CN. Dose response models for infectious gastroenteritis. Risk Anal. 1999;19:1251–60. DOIPubMedGoogle Scholar
- Li Y, Handel A. Modeling inoculum dose dependent patterns of acute virus infections. J Theor Biol. 2014;347:63–73. DOIPubMedGoogle Scholar
- Handel A, Li Y, McKay B, Pawelek KA, Zarnitsyna V, Antia R. Exploring the impact of inoculum dose on host immunity and morbidity to inform model-based vaccine design. PLOS Comput Biol. 2018;14:
e1006505 . DOIPubMedGoogle Scholar - Moore JR, Ahmed H, Manicassamy B, Garcia-Sastre A, Handel A, Antia R. varying inoculum dose to assess the roles of the immune response and target cell depletion by the pathogen in control of acute viral infections. Bull Math Biol. 2020;82:35. DOIPubMedGoogle Scholar
- Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, et al. Determination of the 50% human infectious dose for Norwalk virus. J Infect Dis. 2014;209:1016–22. DOIPubMedGoogle Scholar
- Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, et al. Norwalk virus shedding after experimental human infection. Emerg Infect Dis. 2008;14:1553–7. DOIPubMedGoogle Scholar
- Kavanagh O, Estes MK, Reeck A, Raju RM, Opekun AR, Gilger MA, et al. Serological responses to experimental Norwalk virus infection measured using a quantitative duplex time-resolved fluorescence immunoassay. Clin Vaccine Immunol. 2011;18:1187–90. DOIPubMedGoogle Scholar
- Czakó R, Atmar RL, Opekun AR, Gilger MA, Graham DY, Estes MK. Serum hemagglutination inhibition activity correlates with protection from gastroenteritis in persons infected with Norwalk virus. Clin Vaccine Immunol. 2012;19:284–7. DOIPubMedGoogle Scholar
- Ajami NJ, Barry MA, Carrillo B, Muhaxhiri Z, Neill FH, Prasad BV, et al. Antibody responses to norovirus genogroup GI.1 and GII.4 proteases in volunteers administered Norwalk virus. Clin Vaccine Immunol. 2012;19:1980–3. DOIPubMedGoogle Scholar
- McElreath R. Statistical rethinking: a Bayesian course with examples in R and Stan. 2nd edition. Boca Raton: Taylor and Francis, CRC Press; 2020.
- R Core Team. R: a language and environment for statistical computing. 2017 [cited 2023 Apr 22]. https://www.r-project.org
- Carpenter B, Gelman A, Hoffman MD, Lee D, Goodrich B, Betancourt M, et al. Stan: a probabilistic programming language. J Stat Softw. 2017;76:1–32. DOIPubMedGoogle Scholar
- Burkner P-C. Brms: an R Package for Bayesian multilevel models using Stan. J Stat Softw. 2017;80:80. DOIGoogle Scholar
- Holder BP, Beauchemin CA. Exploring the effect of biological delays in kinetic models of influenza within a host or cell culture. BMC Public Health. 2011;11(Suppl 1):S10. DOIPubMedGoogle Scholar
- Ruuska T, Vesikari T. Rotavirus disease in Finnish children: use of numerical scores for clinical severity of diarrhoeal episodes. Scand J Infect Dis. 1990;22:259–67. DOIPubMedGoogle Scholar
- Freedman SB, Eltorky M, Gorelick M; Pediatric Emergency Research Canada Gastroenteritis Study Group. Evaluation of a gastroenteritis severity score for use in outpatient settings. Pediatrics. 2010;125:e1278–85. DOIPubMedGoogle Scholar
- Bierhoff M, Arvelo W, Estevez A, Bryan J, McCracken JP, López MR, et al. Incidence and clinical profile of norovirus disease in Guatemala, 2008–2013. Clin Infect Dis. 2018;67:430–6. DOIPubMedGoogle Scholar
- Shim DH, Kim DY, Cho KY. Diagnostic value of the Vesikari Scoring System for predicting the viral or bacterial pathogens in pediatric gastroenteritis. Korean J Pediatr. 2016;59:126–31. DOIPubMedGoogle Scholar
- Hornick RB, Greisman SE, Woodward TE, DuPont HL, Dawkins AT, Snyder MJ. Typhoid fever: pathogenesis and immunologic control. N Engl J Med. 1970;283:686–91. DOIPubMedGoogle Scholar
- Hornick RB, Music SI, Wenzel R, Cash R, Libonati JP, Snyder MJ, et al. The Broad Street pump revisited: response of volunteers to ingested cholera vibrios. Bull N Y Acad Med. 1971;47:1181–91.PubMedGoogle Scholar
- Bernstein DI, Atmar RL, Lyon GM, Treanor JJ, Chen WH, Jiang X, et al. Norovirus vaccine against experimental human GII.4 virus illness: a challenge study in healthy adults. J Infect Dis. 2015;211:870–8. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 29, Number 7—July 2023
| EID Search Options |
|---|
|
|
|
|
|
|




Please use the form below to submit correspondence to the authors or contact them at the following address:
Yang Ge, 118 College Dr, School of Health Professions, University of Southern Mississippi, Hattiesburg, MS 39402, USA
Top