Volume 30, Number 8—August 2024
Research
SARS-CoV-2 Seropositivity in Urban Population of Wild Fallow Deer, Dublin, Ireland, 2020–2022
Figure 2
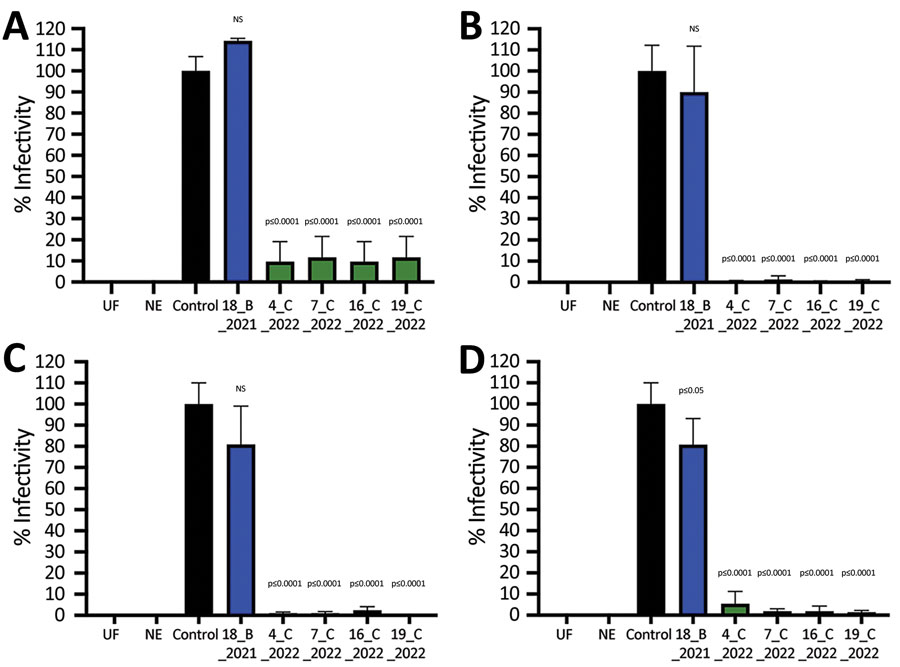
Figure 2. Infectivity of SARS-CoV-2 pseudoviruses after incubation with SARS-CoV-2–positive serum samples from wild fallow deer, Dublin, Ireland, 2020–2022. Spike proteins were from Alpha (A), Delta (B), Omicron BA.1 (C), and Omicron BA.2 (D) variants of concern. SARS-CoV-2 pseudoviruses bearing spike proteins from different variants of concern were incubated with 5 deer serum samples at a 1:1 ratio in triplicate and then used to infect Vero E6/TMPRSS2 cells. Identification numbers of deer are indicated. Controls were virus incubated in triplicate at a 1:1 ratio with culture medium. Relative light units from a luciferase reporter were used to calculate percentage infectivity relative to the untreated control virus. Data are from 2 independent experiments with 3 biologic replicates per experiment. Error bars indicate SDs. NE, no envelope naked pseudovirus control; NS, not significant; UF, uninfected cells.
1These authors contributed equally to this article.