Volume 30, Number 8—August 2024
Research
SARS-CoV-2 Seropositivity in Urban Population of Wild Fallow Deer, Dublin, Ireland, 2020–2022
Figure 9
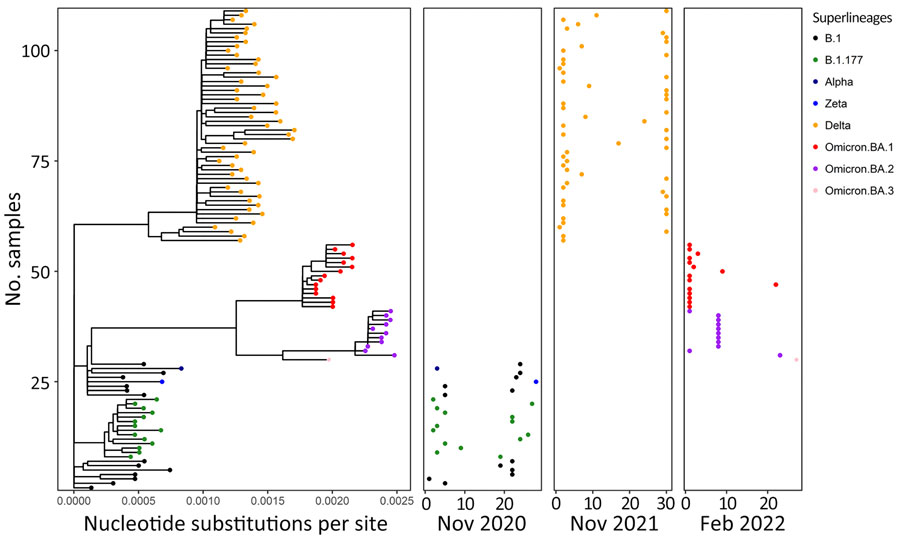
Figure 9. Phylogenetic analysis of SARS-CoV-2 superlineages circulating in humans during deer sampling months in study of SARS-CoV-2 seropositivity in wild fallow deer, Dublin, Ireland, 2020–2022. We analyzed SARS-CoV-2 whole-genome sequences from human clinical samples collected in Ireland covering months corresponding to the deer culling dates (November 2020, November 2021, and February 2022). Branch lengths in the phylogenetic tree (left panel) show the number of base substitutions per site. Colors indicate different SARS-CoV-2 variants. Pangolin lineages are shown with corresponding major circulating variants for each cull month. Location of dots shown for each cull month (right 3 panels) corresponds to the sampling date in each month (horizontal axis) and the phylogenetic position within the tree panel (vertical axis).
1These authors contributed equally to this article.