Volume 31, Number 1—January 2025
Research
Cefiderocol Resistance Conferred by Plasmid-Located Ferric Citrate Transport System in KPC-Producing Klebsiella pneumoniae
Figure 3
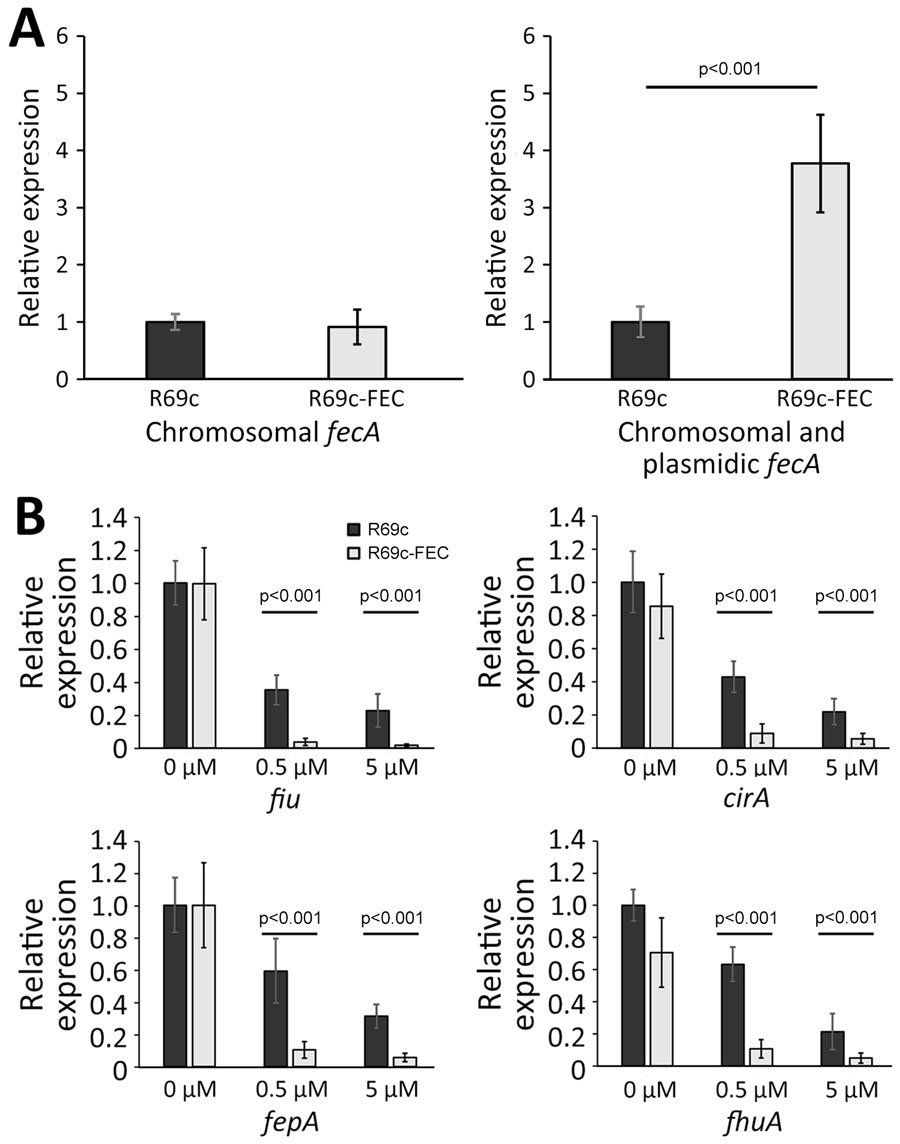
Figure 3. Expression analysis of siderophore receptor genes in the presence and absence of plasmidic fec gene cluster and ferric citric inducer in an Escherichia coli model used in a study of cefiderocol resistance conferred by plasmid-located ferric citrate transport system KPC-producing Klebsiella pneumoniae. A) Transcription of the fecA genes in the DH5-α strain carrying R69c or R69c- FEC, determined by using primer pairs able to discern the chromosomal fecA allele from the K. pneumoniae fecA gene in the fecABCDE operon or a primer pair recognizing both chromosomal and plasmidic fecA alleles (Appendix 1 Table 2). The relative quantitative analysis of the transcripts was based on the 2−ΔΔCT method (31). In both bar graphs, the relative values were calculated with respect to the transcript level observed in the R69c carrying strains and set to 1. B) Transcription of the siderophore receptor genes fiu, cirA, fepA, and fhuA in the R69c-FEC and R69c strains grown in the absence of ferric citrate or in the presence of 0.5 μM or 5.0 μM ferric citrate, relative to the R69c strain grown without ferric citrate, which is set to 1. The relative quantitative analysis of the transcripts was based on the 2−ΔΔCT method (31). Error bars represent SDs. Statistical significance was determined by using a paired 2-tailed Student t-test comparing the dataset obtained from the 2 strains grown under the same conditions. FEC, ferric citrate transport system; KPC, Klebsiella pneumoniae carbapenemase.
References
- Miller WR, Arias CA. ESKAPE pathogens: antimicrobial resistance, epidemiology, clinical impact and therapeutics. Nat Rev Microbiol. 2024;22:598–616. DOIPubMedGoogle Scholar
- World Health Organization. WHO bacterial priority pathogens list, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance [cited 2024 Nov 27]. https://www.who.int/publications/i/item/9789240093461
- Tamma PD, Han JH, Rock C, Harris AD, Lautenbach E, Hsu AJ, et al.; Antibacterial Resistance Leadership Group. Carbapenem therapy is associated with improved survival compared with piperacillin-tazobactam for patients with extended-spectrum β-lactamase bacteremia. Clin Infect Dis. 2015;60:1319–25. DOIPubMedGoogle Scholar
- Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59:5873–84. DOIPubMedGoogle Scholar
- Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–61. DOIPubMedGoogle Scholar
- Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–96. DOIPubMedGoogle Scholar
- Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J Antimicrob Chemother. 2010;65:243–8. DOIPubMedGoogle Scholar
- Shirley M. Ceftazidime-avibactam: a review in the treatment of serious gram-negative bacterial infections. Drugs. 2018;78:675–92. DOIPubMedGoogle Scholar
- Yahav D, Giske CG, Grāmatniece A, Abodakpi H, Tam VH, Leibovici L. New β-lactam-β-lactamase inhibitor combinations. Clin Microbiol Rev. 2020;34:e00115–20. DOIPubMedGoogle Scholar
- Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, et al. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother. 2017;61:e02097–16.PubMedGoogle Scholar
- Humphries RM, Yang S, Hemarajata P, Ward KW, Hindler JA, Miller SA, et al. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother. 2015;59:6605–7. DOIPubMedGoogle Scholar
- Hobson CA, Pierrat G, Tenaillon O, Bonacorsi S, Bercot B, Jaouen E, et al. Klebsiella pneumoniae carbapenemase variants resistant to ceftazidime-avibactam: an evolutionary overview. Antimicrob Agents Chemother. 2022;66:
e0044722 . DOIPubMedGoogle Scholar - Taracila MA, Bethel CR, Hujer AM, Papp-Wallace KM, Barnes MD, Rutter JD, et al. Different conformations revealed by NMR underlie resistance to ceftazidime/avibactam and susceptibility to meropenem and imipenem among D179Y variants of KPC β-lactamase. Antimicrob Agents Chemother. 2022;66:
e0212421 . DOIPubMedGoogle Scholar - Sato T, Yamawaki K. Cefiderocol: discovery, chemistry, and in vivo profiles of a novel siderophore cephalosporin. Clin Infect Dis. 2019;69(Suppl 7):S538–43. DOIPubMedGoogle Scholar
- Wu JY, Srinivas P, Pogue JM. Cefiderocol: a novel agent for the management of multidrug-resistant gram-negative organisms. Infect Dis Ther. 2020;9:17–40. DOIPubMedGoogle Scholar
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Freire B, Ladra S, Parama JR. Memory-efficient assembly using Flye. IEEE/ACM Trans Comput Biol Bioinform. 2022;19:3564–77. DOIPubMedGoogle Scholar
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:
e1005595 . DOIPubMedGoogle Scholar - Bouras G, Houtak G, Wick RR, Mallawaarachchi V, Roach MJ, Papudeshi B, et al. Hybracter: enabling scalable, automated, complete and accurate bacterial genome assemblies. Microb Genom. 2024;10:
001244 . DOIPubMedGoogle Scholar - Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- von Gabain A, Belasco JG, Schottel JL, Chang AC, Cohen SN. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A. 1983;80:653–7. DOIPubMedGoogle Scholar
- Trirocco R, Pasqua M, Tramonti A, Grossi M, Colonna B, Paiardini A, et al. Fatty acids abolish Shigella virulence by inhibiting its master regulator, VirF. Microbiol Spectr. 2023;11:
e0077823 . DOIPubMedGoogle Scholar - Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. DOIPubMedGoogle Scholar
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. DOIPubMedGoogle Scholar
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar
- Carattoli A, Arcari G, Bibbolino G, Sacco F, Tomolillo D, Di Lella FM, et al. Evolutionary trajectories toward ceftazidime-avibactam resistance in Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2021;65:
e0057421 . DOIPubMedGoogle Scholar - Arcari G, Cecilia F, Oliva A, Polani R, Raponi G, Sacco F, et al. Genotypic evolution of Klebsiella pneumoniae sequence type 512 during ceftazidime/avibactam, meropenem/vaborbactam, and cefiderocol treatment, Italy. Emerg Infect Dis. 2023;29:2266–74. DOIPubMedGoogle Scholar
- Braun V, Hartmann MD, Hantke K. Transcription regulation of iron carrier transport genes by ECF sigma factors through signaling from the cell surface into the cytoplasm. FEMS Microbiol Rev. 2022;46:
fuac010 . DOIPubMedGoogle Scholar - Villa L, Feudi C, Fortini D, Brisse S, Passet V, Bonura C, et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom. 2017;3:
e000110 . DOIPubMedGoogle Scholar - Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25:402–8. DOIPubMedGoogle Scholar
- Enz S, Mahren S, Menzel C, Braun V. Analysis of the ferric citrate transport gene promoter of Escherichia coli. J Bacteriol. 2003;185:2387–91. DOIPubMedGoogle Scholar
- Angerer A, Braun V. Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch Microbiol. 1998;169:483–90. DOIPubMedGoogle Scholar
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. DOIPubMedGoogle Scholar
- Simner PJ, Bergman Y, Conzemius R, Jacobs E, Tekle T, Beisken S, et al. An NDM-producing Escherichia coli clinical isolate exhibiting resistance to cefiderocol and the combination of ceftazidime-avibactam and aztreonam: another step toward pan-β-lactam resistance. Open Forum Infect Dis. 2023;10:
ofad276 . DOIPubMedGoogle Scholar - McElheny CL, Fowler EL, Iovleva A, Shields RK, Doi Y. In vitro evolution of cefiderocol resistance in an NDM-producing Klebsiella pneumoniae due to functional loss of CirA. Microbiol Spectr. 2021;9:
e0177921 . DOIPubMedGoogle Scholar - Lan P, Lu Y, Jiang Y, Wu X, Yu Y, Zhou J. Catecholate siderophore receptor CirA impacts cefiderocol susceptibility in Klebsiella pneumoniae. Int J Antimicrob Agents. 2022;60:
106646 . DOIPubMedGoogle Scholar - Poirel L, Sadek M, Kusaksizoglu A, Nordmann P. Co-resistance to ceftazidime-avibactam and cefiderocol in clinical isolates producing KPC variants. Eur J Clin Microbiol Infect Dis. 2022;41:677–80. DOIPubMedGoogle Scholar
- Coppi M, Di Pilato V, Monaco F, Giani T, Conaldi PG, Rossolini GM. Ceftazidime-avibactam resistance associated with increased blaKPC-3 gene copy number mediated by pKpQIL plasmid derivatives in sequence type 258 Klebsiella pneumoniae. Antimicrob Agents Chemother. 2020;64:e01816–9. DOIPubMedGoogle Scholar
- Hobson CA, Cointe A, Jacquier H, Choudhury A, Magnan M, Courroux C, et al. Cross-resistance to cefiderocol and ceftazidime-avibactam in KPC β-lactamase mutants and the inoculum effect. Clin Microbiol Infect. 2021;27:1172.e7–10. DOIPubMedGoogle Scholar
- Pressler U, Staudenmaier H, Zimmermann L, Braun V. Genetics of the iron dicitrate transport system of Escherichia coli. J Bacteriol. 1988;170:2716–24. DOIPubMedGoogle Scholar
- Staudenmaier H, Van Hove B, Yaraghi Z, Braun V. Nucleotide sequences of the fecBCDE genes and locations of the proteins suggest a periplasmic-binding-protein-dependent transport mechanism for iron(III) dicitrate in Escherichia coli. J Bacteriol. 1989;171:2626–33. DOIPubMedGoogle Scholar
- Baichoo N, Helmann JD. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol. 2002;184:5826–32. DOIPubMedGoogle Scholar
- Kocer K, Boutin S, Heeg K, Nurjadi D. The acquisition of transferable extrachromosomal fec operon is associated with a cefiderocol MIC increase in Enterobacterales. J Antimicrob Chemother. 2022;77:3487–95. DOIPubMedGoogle Scholar
- Ganz T, Nemeth E. Iron homeostasis in host defence and inflammation. Nat Rev Immunol. 2015;15:500–10. DOIPubMedGoogle Scholar
- Page MGP. The role of iron and siderophores in infection, and the development of siderophore antibiotics. Clin Infect Dis. 2019;69(Suppl 7):S529–37. DOIPubMedGoogle Scholar
- Costello LC, Franklin RB. Plasma citrate homeostasis: how it is regulated; and its physiological and clinical implications. an important, but neglected, relationship in medicine. HSOA J Hum Endocrinol. 2016;1:005.
- Frick-Cheng AE, Sintsova A, Smith SN, Pirani A, Snitkin ES, Mobley HLT. Ferric citrate uptake is a virulence factor in uropathogenic Escherichia coli. MBio. 2022;13:
e0103522 . DOIPubMedGoogle Scholar - Huang WC, Wong MY, Wang SH, Hashimoto M, Lin MH, Lee MF, et al. The ferric citrate uptake system encoded in a novel bla CTX-M-3- and bla TEM-1-harboring conjugative plasmid contributes to the virulence of Escherichia coli. Front Microbiol. 2021;12:
667782 . DOIPubMedGoogle Scholar - Blum SE, Goldstone RJ, Connolly JPR, Répérant-Ferter M, Germon P, Inglis NF, et al. Postgenomics characterization of an essential genetic determinant of mammary pathogenic Escherichia coli. MBio. 2018;9:e00423–18. DOIPubMedGoogle Scholar