Volume 31, Number 1—January 2025
Research
Cefiderocol Resistance Conferred by Plasmid-Located Ferric Citrate Transport System in KPC-Producing Klebsiella pneumoniae
Figure 5
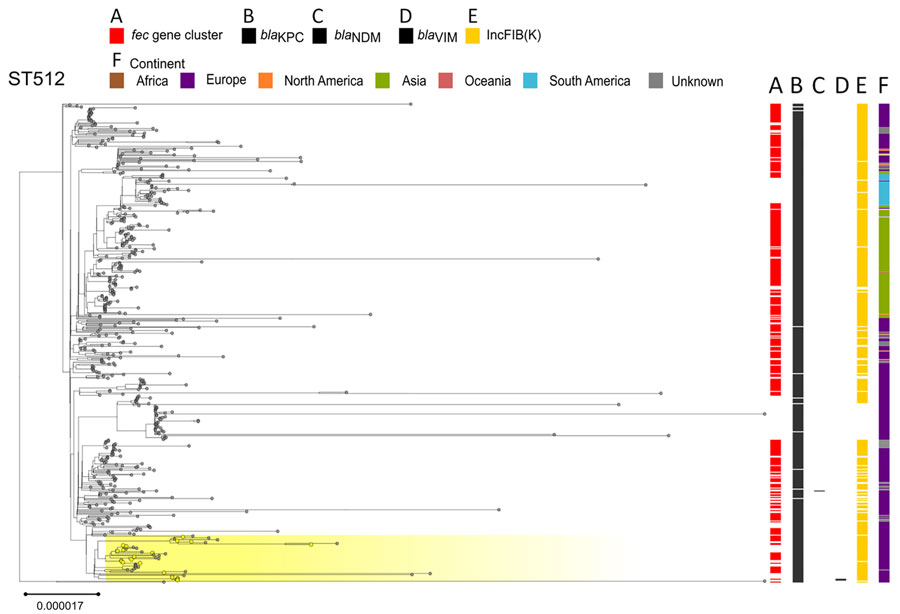
Figure 5. Phylogenetic analysis of Klebsiella pneumoniae ST512 based on core-genome alignment of 510 K. pneumoniae ST512 isolates. The tree is midpoint rooted, and the scale bar represents the number of substitutions per site. The presence of the fec operon is indicated in red; blaKPC, blaVIM, and blaNDM genes in black; and the FIB(K) replicon in orange. Yellow shading indicates genomes sequenced in this study or our previous studies (Appendix 1, Table 1). The best-fit model was selected by ModelFinder (http://www.iqtree.org/ModelFinder), The tree was visualized with Microreact (https://microreact.org) and adjusted by using the InkScape software (https://www.inkscape.org). ST, sequence type.
Page created: December 02, 2024
Page updated: December 22, 2024
Page reviewed: December 22, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.