Volume 31, Number 1—January 2025
Research
Cefiderocol Resistance Conferred by Plasmid-Located Ferric Citrate Transport System in KPC-Producing Klebsiella pneumoniae
Abstract
Cefiderocol (FDC), a siderophore-cephalosporin conjugate, is the newest option for treating infection with carbapenem-resistant gram-negative bacteria. We identified a novel mechanism contributing to decreased FDC susceptibility in Klebsiella pneumoniae clinical isolates. The mechanism involves 2 coresident plasmids: pKpQIL, carrying variants of blaKPC carbapenemase gene, and pKPN, carrying the ferric citrate transport (FEC) system. We observed increasing FDC MICs in an Escherichia coli model system carrying different natural pKpQIL plasmids, encoding different K. pneumoniae carbapenemase (KPC) variants, in combination with a conjugative low copy number vector carrying the fec gene cluster from pKPN. We observed transcriptional repression of fiu, cirA, fepA, and fhuA siderophore receptor genes in blaKPC–fec–E. coli cells treated with ferric citrate. Screening of 27,793 K. pneumoniae whole-genome sequences revealed that the fec cluster occurs frequently in some globally distributed different KPC-producing K. pneumoniae clones (sequence types 258, 14, 45, and 512), contributing to reduced FDC susceptibility.
Klebsiella pneumoniae is 1 of 6 global leading ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) associated with high morbidity and mortality rates and antimicrobial resistance (1). Among those pathogens, the World Health Organization designated carbapenem-resistant K. pneumoniae as a priority pathogen (2). In 1996, K. pneumoniae carbapenemase (KPC) was identified in K. pneumoniae sequence type (ST) 258 in the United States and then spread worldwide (3–5). To date, blaKPC gene variants have been found on different plasmid types; pKpQIL is prevalent in successful clones ST258 and ST512 (6,7).
Since 2015, ceftazidime/avibactam (CZA) has been available for treatment of complicated and deep-seated infections, including bacteremia caused by carbapenemase-producing Enterobacterales in adults (8). CZA combines ceftazidime, a third-generation cephalosporin, and avibactam, a β-lactamase inhibitor (9). Since 2018, KPC-producing CZA-resistant K. pneumoniae strains have been described (10,11) carrying mutations in the Ω-loop of the KPC protein, such as KPC-31 (12,13).
Cefiderocol (FDC), approved by the US Food and Drug Administration in 2020, is available to treat Enterobacterales, Acinetobacter baumannii, and Pseudomonas aeruginosa invasive infections caused by carbapenem- and CZA-resistant strains (https://www.accessdata.fda.gov/drugsatfda_docs/nda/2020/209445Orig1s002.pdf). FDC is a cephalosporin linked with a chlorocatechol group, which provides the drug with a siderophore-like moiety that serves as a Trojan horse to gain access to the bacterial periplasm. The chlorocatechol group is thought to enhance the entry of FDC in the bacterial cell through energy-dependent uptake by chromosome-encoded ferric siderophore transporters (14,15). To determine the mechanism of FDC resistance in KPC-producing K. pneumoniae, we analyzed the contribution of a plasmid-encoded ferric citrate uptake system (FEC), which acts synergistically with CZA-resistant KPC variants.
K. pneumoniae Isolates
K. pneumoniae strains PL1, PL2, PL3, and PL4 were isolated at the University Hospital Policlinico Umberto I, Rome, Italy, from blood samples of 1 patient during 1 month of hospitalization. The strains were processed for routine diagnostics and compared with 31 previously described strains (Appendix 1, Table 1).
FDC Antimicrobial Susceptibility
We determined FDC MICs by using ComASP (Liofilchem, https://www.liofilchem.com) or by using a ComASP panel enriching iron-depleted cation-adjusted Mueller-Hinton broth with 0.5 μM or 5.0 µM ammonium ferric citrate or trisodium citrate dihydrate (Merck KGaA, https://www.emdgroup.com). We preliminarily determined the citrate concentration used for induction of the FEC system in the E. coli model, and 0.5 µM ammonium ferric citrate was the minimal concentration that did not increase the FDC MIC by >1-fold in treated compared with untreated E. coli strains.
Whole-Genome Sequencing
We purified genomic DNA by using the MagaBio Bacterium DNA Purification Kit III (Hangzhou Bioer Technology Co., https://www.bioer.com) and GenePure Pro Nucleic Acid Purification System (Bioer Technology, https://www.bioer.com.cn), and we used NanoDrop One Microvolume UV-Vis Spectrophotometer and Qubit 4.0 Fluorometer (Invitrogen, https://www.thermofisher.com) to assess. We prepared DNA libraries by using the Nextera XT DNA Library Preparation Kit and loaded them onto a MiSeq Reagent Kit v.3 cartridge (Illumina, https://www.illumina.com). We performed paired-end sequencing on an Illumina MiSeq platform, with a read length of 2 × 300 bp. We trimmed resulting reads by using trimmomatic (16) and assembled them by using SPAdes (17).
Long-Reads Sequencing
We performed Oxford Nanopore Technologies (ONT) sequencing on a MinION Mk1C sequencing platform (https://nanoporetech.com). We extracted genomic DNA by using a Monarch HMW DNA Extraction Kit for Tissue (NEB, https://www.neb.com) and prepared libraries by using ONT Rapid Barcoding Kit 24 and sequencing on R10.4.1 flow cells. We performed long-read assemblies by using Flye (18).
We analyzed hybrid assembly obtained by Unicycler (19) and Hybracter (20) by using Staramr (https://bio.tools/staramr). We annotated genomes by using Prokka (21) and identified single-nucleotide polymorphisms by using Snippy (https://github.com/tseemann/snippy).
pKpQIL Plasmid Transformation
We introduced plasmid DNA extracted by using a Pureyield Plasmid Midiprep System (Promega, https://www.promega.com) in chemically competent E. coli Max efficiency DH5-α cells (Thermo Fisher Scientific, https://www.thermofisher.com). We selected transformants on Luria broth (LB) agar plates containing ceftazidime (6 mg/L).
R69c and R69c-FEC Plasmid Assembly
We obtained the R69c vector (GenBank accession no. PQ130559) by cloning the chloramphenicol resistance catA gene amplified from Addgene plasmid #46569 in the SmaI site of R69#1 (European patent EP3541942B1, A. Carattoli, A. Endimiani, https://patents.google.com/patent/EP3541942B1/de?oq=EP3541942B1, accessed November 29, 2024). We obtained PCR products by using PCRBIO VeriFi Polymerase (PCR Biosystems, https://pcrbio.com) and primers listed in Appendix 1, Table 2.
We obtained the R69c-FEC vector (GenBank accession no. PQ085644) by cloning the 7,993-bp fec PCR product (Appendix 1, Figure 1) from the PL3 strain and cloned it in R69c PmeI site. Both clonings were performed by using GeneArt Gibson Assembly HiFi Master Mix (Thermo Fisher Scientific).
We extracted plasmids by using ZymoPURE II Plasmid Maxiprep Kit (Zymo Research, https://www.zymoresearch.com) concentrated by Microcon DNA Fast Flow device (Merck KGaA). We performed transformations by using MAX Efficiency DH5α-T1R Competent Cells (Thermo Fisher Scientific) selected on chloramphenicol 25 mg/L LB agar plates.
Plasmid Conjugation
We grew donor and recipient strains separately in LB broth without antibiotics at an optical density of 1 McFarland. We pooled 50 μL of each culture and dropped 20 μL of the mixture on an LB plate without antibiotics and incubated it at 37°C for 6–10 hours. A patina from the conjugation spot was diluted and plated on LB agar plates containing 25 mg/L chloramphenicol and 6 mg/L CAZ. We sequenced selected positive exconjugants by using Illumina and ONT procedures.
RNA Isolation and Quantitative Reverse Transcription PCR
We conducted bacterial RNA purification on R69c/R69c-FEC-PL3 exconjugant pairs grown in liquid iron-depleted media in the presence of 0 μM, 0.5 μM, or 5.0 μM ammonium ferric citrate by using a hot phenol extraction method (22). We conducted cDNA synthesis and quantitative reverse transcription PCR analyses on a 7300 real-time PCR system (Applied Biosystems, https://www.thermofisher.com) (23). We obtained cDNA for nusA (used as normalizer), fiu, cirA, fepA, fhuA, or fecA genes (Appendix 1, Table 2).
Global Distribution of fec Gene Cluster
As of July 4, 2024, we downloaded 27,993 K. pneumoniae genomes from the Pasteur Institute BIGSdb Klebsiella pneumoniae database (https://bigsdb.pasteur.fr) together with their metadata (Appendix 2, Table). We included 35 strains from our study and previous studies in the collection (Appendix 1, Table 1).
We screened K. pneumoniae genomes for the fec operon by using the BLASTN tool in the Bacterial Isolate Genome Sequence BIGSdb database (https://bigsdb.pasteur.fr). We used as reference the fecABCDE operon and fecIR regulatory genes from K. pneumoniae PL3 (GenBank accession no. CP168103, nt positions 113339–121331). The fec gene cluster was considered present if the E-value was <1e−10 and identity was >85% across >90% of the sequence length.
We assessed the presence of the FIB(K) replicon (GenBank accession no. JN233704) by using the BLASTN tool. We also determined the presence of blaVIM, blaNDM, and blaKPC genes by using the gene presence tool at the Pasteur Institute website (https://bigsdb.pasteur.fr) with minimum percentage identity of 95% and a minimum percentage alignment of 99%.
Phylogenetic Analysis
We used the Prokka tool (20) to annotate 2,493 genomes belonging to ST101, ST307, and ST512 (Appendix 3, Table). We used Roary (24) to generate core-genome alignments, using MAFFT (25), accordingly, to the ST and IQ-TREE 2 (26) to construct phylogenetic trees with 1,000 ultrafast bootstrap iterations.
Effect of KPC Variants on Reduced Susceptibility to FDC
Our initial aim with this study was to explain the different levels of FDC resistance in 4 ST512 K. pneumoniae clinical isolates (PL1, PL2, PL3, and PL4) from 1 patient during 1 month of hospitalization. PL3 was resistant to FDC (MIC 4 mg/L), whereas FDC MICs PL1, PL2, and PL4 were below the breakpoint value (0.5–2.0 mg/L; Appendix 1, Table 1). The PL1–4 genomes were closely related at the chromosomal level (6–22 single-nucleotide polymorphisms and indels on the chromosome; Appendix 4, Table) but showed different blaKPC genes and plasmid content. All PL1–4 K. pneumoniae strains carried different pKpQIL variants plus an IncX3 plasmid and a small ColRNAI plasmid. The pKpQIL-PL1 plasmid harbored 2 copies of the blaKPC-3 gene, both pKpQIL-PL2 and pKpQIL-PL4 carried 1 copy of blaKPC-3 and 1 copy of blaKPC-31, whereas pKpQIL-PL3 carried 2 copies of blaKPC-31 (Appendix 1, Figure 2). Isolates PL2 and PL3 were also enriched with the pKPN plasmid, which was absent in PL1 and PL4. In PL2, pKPN was fused with pKpQIL-PL2 in the tnpR-FIIK1 integration site, forming a 263,486-bp plasmid. The hybrid pKPN-pKpQIL-PL2 plasmid was not transferable by transformation or conjugation and could not be further studied. In PL3, the stand-alone pKPN plasmid had acquired the fec gene cluster encoding for a FEC system. The fec gene cluster was unique to the pKPN plasmid in PL3 and was absent in PL1, PL2, and PL4. The fec genes mapped (alongside an ABC glutathione transporter, the lacZ, lacY, and lacI genes) between 2 IS4321 elements positioned between the tnpR gene and the FIIK replicon (Appendix 1, Figure 3).
Acquisition of fec genes was suspected to correlate with increased FDC MICs of PL3. We then measured FDC MICs for all KPC-producing K. pneumoniae clinical strains in our collection isolated since 2018 with a completely sequenced genome, in search for fec, other siderophore receptors, and porin gene sequences in their genomes (Appendix 1, Table 1).
FDC MICs were 0.25–32 mg/L. The lowest (0.25 mg/L) was measured in a strain producing KPC-3 (strain 3), encoding the yersiniabactin siderophore-dependent iron uptake system, the wild-type CirA and Fiu siderophore receptors, and a wild-type OmpK36 porin (27). The highest FDC MIC (32 mg/L) was for a strain producing VIM (Verona integron-encoded metallo-β-lactamase) and KPC carbapenemases (strain 0296), characterized by a nonsense mutation in the gene encoding the siderophore receptor CirA (E133X) (28).
Presence of CZA-resistant variant KPC-31, KPC-70, or KPC-68 was associated with high MICs (4 mg/L). Higher MICs (1–2 mg/L) for FDC were obtained in E. coli Top-10 transformed with the blaKPC-31, blaKPC-70, or blaKPC-68 genes, respectively cloned in the pTopo-KanR vector (Appendix 1, Table 1).
In addition to the role of KPC variants in determining FDC resistance levels, we noticed that K. pneumoniae exhibiting higher FDC MICs were positive for the FEC system (29), carried by the pKPN plasmid (30). The fec gene cluster was identified in 13/35 isolates from the collection. Eleven KPC-31–producing strains belonging to different STs showed FDC MICs of 1–2 mg/L, but 2 KPC-31 strains carrying the fec gene cluster reached a MIC of 4.0 mg/L (Appendix 1, Table 1). We hypothesized that the plasmid-borne FEC system could reduce the susceptibility to FDC in K. pneumoniae clinical isolates.
E. coli Model
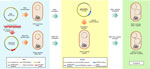
We constructed an in vitro model in isogenic E. coli DH5-α cells, suitable for studying the effect of the FEC transport system on FDC resistance levels, excluding the contribution of other resistance factors, siderophore receptors, and porins encoded by the K. pneumoniae clinical isolates. The model consisted of the 2-step introduction in E. coli DH5-α cell of pKpQIL plasmids carrying different KPC variants and an engineered 64-Kb R69c self-conjugative plasmid vector carrying the K. pneumoniae fec gene cluster.
First, selected pKpQIL plasmid variants were individually introduced by transformation into chemically competent E. coli DH5-α cells. We tested the model on pKpQIL transformants obtained from strains 3, 42B, and 1021, encoding the KPC-3, KPC-70, and KPC-31 variants, respectively (Appendix 1, Table 3). We also studied pKpQIL transformants carrying copies of the blaKPC gene obtained from K. pneumoniae PL1, PL3, and PL4 strains.
Second, the 64-Kb R69c, self-conjugative plasmid vector was engineered to host fec genes. The R69c vector is a derivative of the R69 IncM natural plasmid (GenBank accession no. KM406488) and carries the catA gene, conferring chloramphenicol resistance. R69c is a self-conjugative, low-copy-number plasmid that simulates the horizontal transmission of the pKPN natural plasmid. It carries all the genes enabling conjugation at high efficiency (1×10−2 conjugants/recipient cell), conferring stabilization, and the IncM replicon for replication and copy number control (Appendix 1, Figure 1). The plasmid enables cloning and transfer of genetic determinants at low copy numbers by conjugation. Because R69c contains a stabilization system, after chloramphenicol selection of transconjugants, the recipient E. coli clones do not need further antimicrobial selection to ensure plasmid maintenance. The fecIR-fecABCDE gene cluster, including the Fur and iron-regulated promoter regions (32,33) (Appendix 1, Figure 4), were amplifiedcfrom the pKPN-PL3 K. pneumoniae plasmid. The re- sulting PCR product of 7,993 bp, consisting of the fecIR promoter region, the fecI and fecR regulatory genes, and the fecA, fecB, fecC, fecD, and fecE genes encoding the complete FEC system with internal regulatory regions, was cloned in the unique PmeI restriction site of R69c, obtaining the 73 Kb vector, named R69c-FEC (Appendix 1, Figure 1).
Evaluation of FDC MICs in the E. coli Model
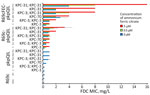
We introduced R69c and R69c-FEC vectors by conjugation in DH5-α pKpQIL transformants (Appendix 1, Table 3) by obtaining pairs of exconjugants carrying the same pKpQIL variant and, alternatively, the R69c vector with or without the cloned fec gene cluster (Figure 1). As an E. coli K-12 derivative, strain DH5-α cells possess the chromosomal fec gene cluster (94.51% nt identity, 96% coverage; Appendix 1, Figure 4). Higher FDC MICs were invariably observed for exconjugants carrying the R69c-FEC, relative to the isogenic strain carrying the same pKpQIL with R69c lacking the fec gene cluster (Figure 2). The highest FDC MIC of 4 mg/L in iron-depleted media was obtained for the exconjugant carrying both pKpQIL-PL3 and R69c-FEC. The respective comparative exconjugant carrying the R69c reached an FDC MIC of 1 mg/L.
We tested in vitro susceptibility to FDC of R69c and R69c-FEC exconjugant pairs under inducing conditions by adding ferric citrate, which serves as substrate and inducer of the FEC system. In our experimental conditions, 0.5 μM or 5.0 μM of ferric citrate increased the FDC MICs of R69c/R69c-FEC DH5-α cells relative to untreated cells. The effect in strains carrying the R69c vector without the cloned fec gene cluster was attributed to the chromosomal fec gene cluster in the DH5-α background. We did not observe increased FDC MICs when using 0 μM, 0.5 μM, and 5.0 μM trisodium citrate dihydrate without iron (data not shown). The highest FDC MICs were reached by R69c-FEC-PL3 (MICs 8 in the presence of 0.5 μM and 16 mg/L in the presence of 5.0 μM ferric citrate). Under the same conditions, R69c-PL3 FDC MICs were 2 and 4 mg/L (Figure 2; Appendix 1, Table 3). The experiments performed with R69c-FEC/R69c-1021, R69c-FEC/R69c-42B, and R69c-FEC/R69c-PL4 pairs (Appendix 1, Table 3) demonstrated that the presence of an inhibitor-resistant KPC variant, such as KPC-31 and KPC-70 in combination with the plasmid-located fec gene cluster was sufficient to reduce FDC susceptibility (MIC >2.0 mg/L) relative to the R69c controls lacking the fec gene cluster. Further FDC MIC increment can be obtained by treatment with ferric citrate (MIC≥4.0 mg/L).
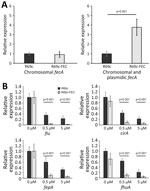
Because the FEC system is implicated in iron delivery to the cell, we compared the mRNA transcription of fiu, cirA, fepA, and fhuA siderophore receptor genes and the endogenous fec gene cluster in DH5-α carrying the R69c-FEC plasmid with DH5-α carrying the R69c, both in the presence or absence of ferric citrate. The expression of the R69c-located fecA cluster was estimated to be 3-fold higher than that of the chromosomal DH5-α fec cluster (Figure 3, panel A).
We observed a substantial reduction of the expression of fiu, cirA, fepA, and fhuA siderophore receptor genes growing the cells in the presence of 0.5 μM and 5.0 μM ferric citrate (relative to no ferric citrate). The inhibition of fiu, cirA, fepA, and fhuA gene expression was almost complete (90%) in cells carrying the R69c-FEC plasmid and only partial in R69c-carrying cells (30%–40%, R69c-FEC vs. R69c; Figure 3, panel B). The markedly reduced expression of ferrisiderophore receptor genes caused by the cloned K. pneumoniae fec gene cluster correlated with the higher FDC MICs observed in R69c-FEC-positive strains in those conditions.
Prevalence of FEC Siderophore Transport System in K. pneumoniae
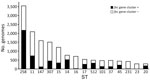
We performed global screening of the fec gene cluster in 27,793 K. pneumoniae whole-genome sequences downloaded from the Pasteur Institute database (https://bigsdb.pasteur.fr/klebsiella; Appendix 2, Table). The resulting dataset included genomes of globally prevalent clones (Figure 4).
The fec gene cluster was detected in 10,672 isolates across the dataset (38.4%), of which 2,658 (24.0%) carried a fec cluster with 100% nt identity and 100% coverage compared with that of K. pneumoniae PL3 (Appendix 5, Table). The fec gene cluster was unevenly spread among the 15 most prevalent STs. The fec gene cluster was carried by a majority (>50%) of ST258, ST14, ST45, and ST512 isolates and a minority (<10%) of ST16, ST147, ST23, and ST231 isolates. The fec genes were more prevalent in ST512 than in the general K. pneumoniae population (68% of fec-carrying ST512 genomes vs. 37.87% of fec-carrying non-ST512 genomes; p<0.00001 by χ2 test). Most ST512 genomes carried 1 copy of the blaKPC carbapenemase gene (Figure 5), whereas the co-presence of fec and blaKPC was less frequent in other clones such as ST101 and ST307 (Figures 6, 7).
Our data suggest that the risk for developing resistance to FDC may be higher in clones like ST512, which more frequently carry the fec gene cluster along with the blaKPC gene. The specific FIB(K) replicon, marking the pKPN plasmid, was detected in 16,325 (58.3%) genomes, of which 2,585 (15.8%) also carried the fec cluster of K. pneumoniae PL3 (Appendix 2, Table).
In vitro studies on FDC resistance have unveiled that CZA-resistant KPC variants (e.g., KPC-31) and class B metallo-β-lactamases have a role in FDC resistance (35). Furthermore, mutational inactivation of CirA and Fiu siderophore receptors has been demonstrated to reduce FDC susceptibility in vitro and in vivo (362,37).
With this study, our first hypothesis was to attribute FDC resistance of PL3 to blaKPC-31 gene duplication (38–40). Subsequently, we noticed that PL3 was unique in carrying the fec gene cluster on the pKPN plasmid compared with the other PL strains. Consequently, acquisition of the fec genes was considered in analysis of genetic traits suspected to increase FDC MIC of PL3. Interest in the FEC transport system was also corroborated by genomic analysis of other K. pneumoniae clinical isolates in our collection, given that isolates showing higher FDC MICs also carried the fec gene cluster.
With regard to the simplified E. coli K-12 laboratory strain model, we speculate that iron imported via the plasmid-encoded FEC system is sufficient to downregulate the expression of Fiu, CirA, FepA and FhuA iron transporters that also mediate FDC import (37,41,42). High intracellular iron levels activate the ferric uptake regulator protein, causing general repression of TonB-dependent transporters (43).
A recent study reported correlation of FDC resistance with a fec gene cluster, originating from E. coli, located on an IncC plasmid in VIM-1–producing Enterobacterales (44). Our findings extend that observation, demonstrating the effect of the widely diffused plasmid-mediated fec gene cluster in globally spread K. pneumoniae KPC-carbapenemase producers. In our model, the combination of 2 plasmids (e.g., pKPN and pKpQIL) resulted in reduced susceptibility to FDC. pKpQIL is one of the most diffused plasmids carrying blaKPC gene variants. pKPN is a plasmid that seems to be restricted to K. pneumoniae and was initially recognized as a vehicle of the fec gene cluster in ST307 (30). We show that the fec gene cluster is present in many K. pneumoniae strains, including those isolated before introduction of FDC in clinical therapy.
Our most relevant evidence is that FDC resistance can be driven by genetic determinants located on plasmids, the success and spread of which occurred independently from the introduction of FDC for therapy. In the future, FDC may act as a positive selector for plasmids carrying the fec genes, for which prevalence can be expected to increase.
The results obtained in the E. coli experimental model should not allow extrapolation or prediction about the clinical efficacy of FDC in treating infections sustained by fec-positive K. pneumoniae. However, our study sets the background for future clinical studies aimed at testing the therapeutic efficacy of FDC in infections caused by K. pneumoniae carrying different combinations of plasmid-encoded carbapenemases and iron-uptake systems.
During infection, bacteria are faced with the low iron availability imposed by the iron-withholding response of the host (45) and must therefore express their iron uptake systems for successful tissue invasion and systemic spread (46). Citrate concentrations in biological fluids (≈100 μM in blood) (47) are high enough to activate the fec gene cluster; accordingly, selective expression of the FEC system has been documented to contribute to in vivo fitness of E. coli in human and animal infection (48–50). Moreover, the introduction of the CZA combination has contributed to selecting K. pneumoniae clinical strains that produce CZA-resistant KPC variants, such as KPC-31, characterized by very efficient cephalosporinase activity on the cephalosporin moiety of FDC (38).
Our novel finding is that the combination of a CZA-resistant KPC variant with the FEC system in K. pneumoniae may substantially increase the FDC MICs. Thus, the Trojan horse approach is a smart and effective strategy for delivering an antimicrobial drug to its target(s), but K. pneumoniae is equipped with plasmids that could help escape that trap.
Dr. Polani is a PhD student in the Life Science PhD School of Sapienza University of Rome, Italy. His main interests are genomics of K. pneumoniae clones and tracing the evolution of mobile genetic elements involved in the spread of antibiotic resistance genes, particularly those conferring resistance to new antibiotics.
Acknowledgments
Whole-genome sequences from this study have been submitted to the National Center for Biotechnology Information under BioProject nos. PRJNA1139702 and PRJNA1139719, and complete plasmid sequences mentioned in the text are under the following GenBank accession nos.: pKpQIL_PL1, accession no. CP168113; pKpQIL_PL3, accession no. CP168102; pKpQIL_PL4, accession no. PQ085643; pKpQIl-pKPN_PL2, accession no. CP168107; plasmid 3-pKpQIL, accession no. ON002623.2; plasmid 42B-pKpQIL, accession no. MT809701; plasmid 17B-pKpQIL, accession no. MT809697; plasmid 1021-pKpQIL, accession no. CP100309; plasmid R69c, accession no. PQ130559; and plasmid R69c FEC, accession no. PQ085644.
This research was supported by EU funding within the NextGeneration EU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases Project no. PE00000007, PE13 INF-ACT to A.C. and L.V. (Spoke 3); Italian Ministry of University and Research PRIN 2022 Project 2022FN7ANE to A.C; PAN_HUB T4-AN-07, Italian Ministry of Health, CUP B83C22004480001 to G. An. P.V. was supported by Rome Technopole, PNRR grant M4-C2-Inv. 1.5, CUPF83B22000040006. R.P. was supported by the PNRR PhD scholarship (ex M.D 351/22) financed by the Rome Technopole Project. A.D.F. was supported by the PNRR PhD scholarship (ex M.D 118/23). PostDoc Fellowship to R.T. was supported by the Istituto Pasteur Italia project 2020 to A.C.
Procedures performed in the study were in accordance with the ethics standards of the Institutional and National Research Committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethics standards.
R.P. and A.D.F. constructed the vectors for the E. coli model, performed transformation and conjugation experiments; R.P. A.D.F., and D.T. analyzed genome and plasmid data, performed phenotypic testing and plasmid assemblies; R.P., I.A., G.Ar., and L.V. contributed to global genomic study; R.P. and M.E. sequenced genomes and completed genome analysis; G.P. and R.T. performed RNA transcript analysis; G.An. coordinated the microbiological work and provided strains; P.V. contributed to project design and data analysis; and A.C. conceived the study, designed and performed data analysis, interpreted results, and wrote the manuscript. All authors contributed to data interpretation and contributed to writing the manuscript.
The authors declare no conflict of interest with respect to the content of this manuscript.
References
- Miller WR, Arias CA. ESKAPE pathogens: antimicrobial resistance, epidemiology, clinical impact and therapeutics. Nat Rev Microbiol. 2024;22:598–616. DOIPubMedGoogle Scholar
- World Health Organization. WHO bacterial priority pathogens list, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance [cited 2024 Nov 27]. https://www.who.int/publications/i/item/9789240093461
- Tamma PD, Han JH, Rock C, Harris AD, Lautenbach E, Hsu AJ, et al.; Antibacterial Resistance Leadership Group. Carbapenem therapy is associated with improved survival compared with piperacillin-tazobactam for patients with extended-spectrum β-lactamase bacteremia. Clin Infect Dis. 2015;60:1319–25. DOIPubMedGoogle Scholar
- Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59:5873–84. DOIPubMedGoogle Scholar
- Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–61. DOIPubMedGoogle Scholar
- Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–96. DOIPubMedGoogle Scholar
- Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J Antimicrob Chemother. 2010;65:243–8. DOIPubMedGoogle Scholar
- Shirley M. Ceftazidime-avibactam: a review in the treatment of serious gram-negative bacterial infections. Drugs. 2018;78:675–92. DOIPubMedGoogle Scholar
- Yahav D, Giske CG, Grāmatniece A, Abodakpi H, Tam VH, Leibovici L. New β-lactam-β-lactamase inhibitor combinations. Clin Microbiol Rev. 2020;34:e00115–20. DOIPubMedGoogle Scholar
- Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, et al. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother. 2017;61:e02097–16.PubMedGoogle Scholar
- Humphries RM, Yang S, Hemarajata P, Ward KW, Hindler JA, Miller SA, et al. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother. 2015;59:6605–7. DOIPubMedGoogle Scholar
- Hobson CA, Pierrat G, Tenaillon O, Bonacorsi S, Bercot B, Jaouen E, et al. Klebsiella pneumoniae carbapenemase variants resistant to ceftazidime-avibactam: an evolutionary overview. Antimicrob Agents Chemother. 2022;66:
e0044722 . DOIPubMedGoogle Scholar - Taracila MA, Bethel CR, Hujer AM, Papp-Wallace KM, Barnes MD, Rutter JD, et al. Different conformations revealed by NMR underlie resistance to ceftazidime/avibactam and susceptibility to meropenem and imipenem among D179Y variants of KPC β-lactamase. Antimicrob Agents Chemother. 2022;66:
e0212421 . DOIPubMedGoogle Scholar - Sato T, Yamawaki K. Cefiderocol: discovery, chemistry, and in vivo profiles of a novel siderophore cephalosporin. Clin Infect Dis. 2019;69(Suppl 7):S538–43. DOIPubMedGoogle Scholar
- Wu JY, Srinivas P, Pogue JM. Cefiderocol: a novel agent for the management of multidrug-resistant gram-negative organisms. Infect Dis Ther. 2020;9:17–40. DOIPubMedGoogle Scholar
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Freire B, Ladra S, Parama JR. Memory-efficient assembly using Flye. IEEE/ACM Trans Comput Biol Bioinform. 2022;19:3564–77. DOIPubMedGoogle Scholar
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:
e1005595 . DOIPubMedGoogle Scholar - Bouras G, Houtak G, Wick RR, Mallawaarachchi V, Roach MJ, Papudeshi B, et al. Hybracter: enabling scalable, automated, complete and accurate bacterial genome assemblies. Microb Genom. 2024;10:
001244 . DOIPubMedGoogle Scholar - Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- von Gabain A, Belasco JG, Schottel JL, Chang AC, Cohen SN. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A. 1983;80:653–7. DOIPubMedGoogle Scholar
- Trirocco R, Pasqua M, Tramonti A, Grossi M, Colonna B, Paiardini A, et al. Fatty acids abolish Shigella virulence by inhibiting its master regulator, VirF. Microbiol Spectr. 2023;11:
e0077823 . DOIPubMedGoogle Scholar - Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. DOIPubMedGoogle Scholar
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. DOIPubMedGoogle Scholar
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar
- Carattoli A, Arcari G, Bibbolino G, Sacco F, Tomolillo D, Di Lella FM, et al. Evolutionary trajectories toward ceftazidime-avibactam resistance in Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2021;65:
e0057421 . DOIPubMedGoogle Scholar - Arcari G, Cecilia F, Oliva A, Polani R, Raponi G, Sacco F, et al. Genotypic evolution of Klebsiella pneumoniae sequence type 512 during ceftazidime/avibactam, meropenem/vaborbactam, and cefiderocol treatment, Italy. Emerg Infect Dis. 2023;29:2266–74. DOIPubMedGoogle Scholar
- Braun V, Hartmann MD, Hantke K. Transcription regulation of iron carrier transport genes by ECF sigma factors through signaling from the cell surface into the cytoplasm. FEMS Microbiol Rev. 2022;46:
fuac010 . DOIPubMedGoogle Scholar - Villa L, Feudi C, Fortini D, Brisse S, Passet V, Bonura C, et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom. 2017;3:
e000110 . DOIPubMedGoogle Scholar - Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25:402–8. DOIPubMedGoogle Scholar
- Enz S, Mahren S, Menzel C, Braun V. Analysis of the ferric citrate transport gene promoter of Escherichia coli. J Bacteriol. 2003;185:2387–91. DOIPubMedGoogle Scholar
- Angerer A, Braun V. Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch Microbiol. 1998;169:483–90. DOIPubMedGoogle Scholar
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. DOIPubMedGoogle Scholar
- Simner PJ, Bergman Y, Conzemius R, Jacobs E, Tekle T, Beisken S, et al. An NDM-producing Escherichia coli clinical isolate exhibiting resistance to cefiderocol and the combination of ceftazidime-avibactam and aztreonam: another step toward pan-β-lactam resistance. Open Forum Infect Dis. 2023;10:
ofad276 . DOIPubMedGoogle Scholar - McElheny CL, Fowler EL, Iovleva A, Shields RK, Doi Y. In vitro evolution of cefiderocol resistance in an NDM-producing Klebsiella pneumoniae due to functional loss of CirA. Microbiol Spectr. 2021;9:
e0177921 . DOIPubMedGoogle Scholar - Lan P, Lu Y, Jiang Y, Wu X, Yu Y, Zhou J. Catecholate siderophore receptor CirA impacts cefiderocol susceptibility in Klebsiella pneumoniae. Int J Antimicrob Agents. 2022;60:
106646 . DOIPubMedGoogle Scholar - Poirel L, Sadek M, Kusaksizoglu A, Nordmann P. Co-resistance to ceftazidime-avibactam and cefiderocol in clinical isolates producing KPC variants. Eur J Clin Microbiol Infect Dis. 2022;41:677–80. DOIPubMedGoogle Scholar
- Coppi M, Di Pilato V, Monaco F, Giani T, Conaldi PG, Rossolini GM. Ceftazidime-avibactam resistance associated with increased blaKPC-3 gene copy number mediated by pKpQIL plasmid derivatives in sequence type 258 Klebsiella pneumoniae. Antimicrob Agents Chemother. 2020;64:e01816–9. DOIPubMedGoogle Scholar
- Hobson CA, Cointe A, Jacquier H, Choudhury A, Magnan M, Courroux C, et al. Cross-resistance to cefiderocol and ceftazidime-avibactam in KPC β-lactamase mutants and the inoculum effect. Clin Microbiol Infect. 2021;27:1172.e7–10. DOIPubMedGoogle Scholar
- Pressler U, Staudenmaier H, Zimmermann L, Braun V. Genetics of the iron dicitrate transport system of Escherichia coli. J Bacteriol. 1988;170:2716–24. DOIPubMedGoogle Scholar
- Staudenmaier H, Van Hove B, Yaraghi Z, Braun V. Nucleotide sequences of the fecBCDE genes and locations of the proteins suggest a periplasmic-binding-protein-dependent transport mechanism for iron(III) dicitrate in Escherichia coli. J Bacteriol. 1989;171:2626–33. DOIPubMedGoogle Scholar
- Baichoo N, Helmann JD. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol. 2002;184:5826–32. DOIPubMedGoogle Scholar
- Kocer K, Boutin S, Heeg K, Nurjadi D. The acquisition of transferable extrachromosomal fec operon is associated with a cefiderocol MIC increase in Enterobacterales. J Antimicrob Chemother. 2022;77:3487–95. DOIPubMedGoogle Scholar
- Ganz T, Nemeth E. Iron homeostasis in host defence and inflammation. Nat Rev Immunol. 2015;15:500–10. DOIPubMedGoogle Scholar
- Page MGP. The role of iron and siderophores in infection, and the development of siderophore antibiotics. Clin Infect Dis. 2019;69(Suppl 7):S529–37. DOIPubMedGoogle Scholar
- Costello LC, Franklin RB. Plasma citrate homeostasis: how it is regulated; and its physiological and clinical implications. an important, but neglected, relationship in medicine. HSOA J Hum Endocrinol. 2016;1:005.
- Frick-Cheng AE, Sintsova A, Smith SN, Pirani A, Snitkin ES, Mobley HLT. Ferric citrate uptake is a virulence factor in uropathogenic Escherichia coli. MBio. 2022;13:
e0103522 . DOIPubMedGoogle Scholar - Huang WC, Wong MY, Wang SH, Hashimoto M, Lin MH, Lee MF, et al. The ferric citrate uptake system encoded in a novel bla CTX-M-3- and bla TEM-1-harboring conjugative plasmid contributes to the virulence of Escherichia coli. Front Microbiol. 2021;12:
667782 . DOIPubMedGoogle Scholar - Blum SE, Goldstone RJ, Connolly JPR, Répérant-Ferter M, Germon P, Inglis NF, et al. Postgenomics characterization of an essential genetic determinant of mammary pathogenic Escherichia coli. MBio. 2018;9:e00423–18. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: December 18, 2024
Table of Contents – Volume 31, Number 1—January 2025
| EID Search Options |
|---|
|
|
|
|
|
|



Please use the form below to submit correspondence to the authors or contact them at the following address:
Alessandra Carattoli, Department of Molecular Medicine, Sapienza University of Rome, Viale Porta Tiburtina 28, 00185 Rome, Italy
Top