Volume 14, Number 7—July 2008
Research
Wide Distribution of a High-Virulence Borrelia burgdorferi Clone in Europe and North America
Figure 2
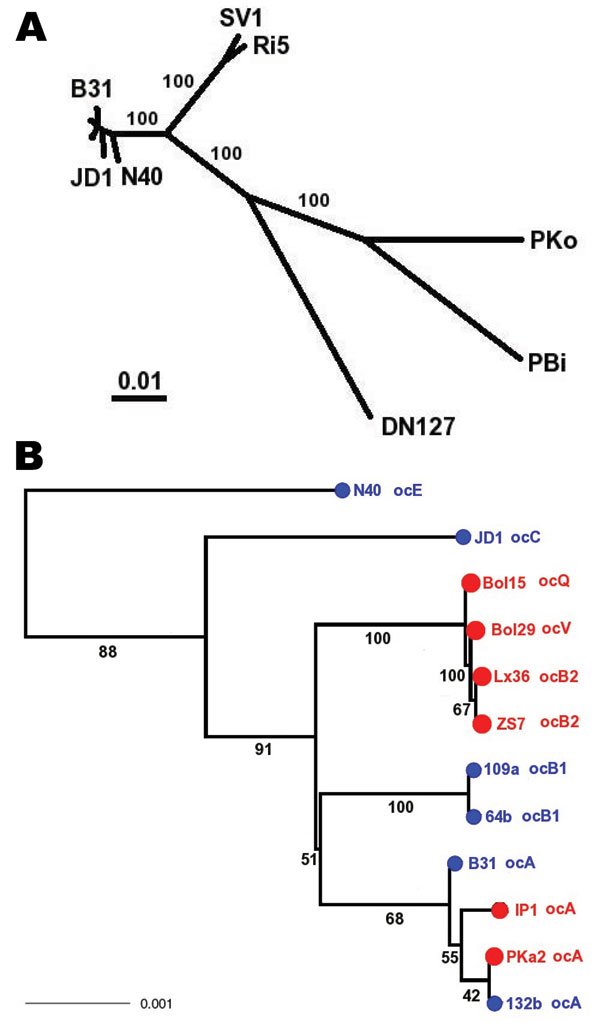
Figure 2. Species phylogeny based on concatenated sequences at housekeeping loci. Seventeen isolates include 1 Borrelia garinii strain (PBi), 1 B. afzelii strain (PKo), 1 B. bissettii strain (DN127), 2 strains of an unnamed genomic species (SV1 and Ri5), and 12 B. burgdorferi sensu stricto isolates. These strains were selected for reconstructing interspecies phylogeny (hence species samples), as well as for resolving the clade containing clonal groups A and B (A, B1, and B2 are represented by 2 isolates). Sequences at 6 chromosomal housekeeping loci (gap, alr, glpA, xylB, ackA, and tgt) were obtained for each strain, with B31 and PBi sequences from published genomes (29,36), N40, JD1, PKo, and DN127 sequences from draft genomes (S. Casjens et al. pers. comm.). Sequences of the remaining strains were obtained by direct sequencing. The total length of concatenated alignment is 7,509 nt. A) Consensus of maximum likelihood trees obtained by using DNAML of the PHYLIP package (33). Branch support values are based on 100 bootstrapped replicates of the original alignment. B) Enlarged view of B. burgdorferi sensu stricto subtree. Tips were colored by geographic origin of the isolate (blue, North America; red, Europe) and were labeled with ospC major-group allele type. Scale bars indicate number of nucleotide substitutions per site.
References
- Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–5. DOIPubMedGoogle Scholar
- Feil EJ. Small change: keeping pace with microevolution. Nat Rev Microbiol. 2004;2:483–95. DOIPubMedGoogle Scholar
- Cohan FM. Concepts of bacterial biodiversity for the age of genomics. In: Fraser CM, Read TD, Nelson KE, editors. Microbial genomes. Totwa (NJ): Humana Press, Inc.; 2004. p. 175–94.
- Fraser C, Hanage WP, Spratt BG. Neutral microepidemic evolution of bacterial pathogens. Proc Natl Acad Sci U S A. 2005;102:1968–73. DOIPubMedGoogle Scholar
- Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Opinion: re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–9. DOIPubMedGoogle Scholar
- Steere AC, Coburn J, Glickstein L. The emergence of Lyme disease. J Clin Invest. 2004;113:1093–101.PubMedGoogle Scholar
- Piesman J, Gern L. Lyme borreliosis in Europe and North America. Parasitology. 2004;129(Suppl):S191–220. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Lyme disease—United States, 2003–2005. MMWR Morb Mortal Wkly Rep. 2007;56:573–6.PubMedGoogle Scholar
- Brisson D, Dykhuizen DE. ospC diversity in Borrelia burgdorferi: different hosts are different niches. Genetics. 2004;168:713–22. DOIPubMedGoogle Scholar
- Hanincova K, Kurtenbach K, Diuk-Wasser M, Brei B, Fish D. Epidemic spread of Lyme borreliosis, northeastern United States. Emerg Infect Dis. 2006;12:604–11.PubMedGoogle Scholar
- Rauter C, Hartung T. Prevalence of Borrelia burgdorferi sensu lato genospecies in Ixodes ricinus ticks in Europe: a metaanalysis. Appl Environ Microbiol. 2005;71:7203–16. DOIPubMedGoogle Scholar
- Gern L, Estrada-Pena A, Frandsen F, Gray JS, Jaenson TG, Jongejan F, European reservoir hosts of Borrelia burgdorferi sensu lato. Zentralbl Bakteriol. 1998;287:196–204.PubMedGoogle Scholar
- Wang IN, Dykhuizen DE, Qiu W, Dunn JJ, Bosler EM, Luft BJ. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics. 1999;151:15–30.PubMedGoogle Scholar
- Alghaferi MY, Anderson JM, Park J, Auwaerter PG, Aucott JN, Norris DE, Borrelia burgdorferi ospC heterogeneity among human and murine isolates from a defined region of northern Maryland and southern Pennsylvania: lack of correlation with invasive and noninvasive genotypes. J Clin Microbiol. 2005;43:1879–84. DOIPubMedGoogle Scholar
- Qiu WG, Dykhuizen DE, Acosta MS, Luft BJ. Geographic uniformity of the Lyme disease spirochete (Borrelia burgdorferi) and its shared history with tick vector (Ixodes scapularis) in the northeastern United States. Genetics. 2002;160:833–49.PubMedGoogle Scholar
- Bunikis J, Garpmo U, Tsao J, Berglund J, Fish D, Barbour AG. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology. 2004;150:1741–55. DOIPubMedGoogle Scholar
- Qiu WG, Schutzer SE, Bruno JF, Attie O, Xu Y, Dunn JJ, Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing. Proc Natl Acad Sci U S A. 2004;101:14150–5. DOIPubMedGoogle Scholar
- Stevenson B, Miller JC. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J Mol Evol. 2003;57:309–24. DOIPubMedGoogle Scholar
- Attie O, Bruno JF, Xu Y, Qiu D, Luft BJ, Qiu WG. Co-evolution of the outer surface protein C gene (ospC) and intraspecific lineages of Borrelia burgdorferi sensu stricto in the northeastern United States. Infect Genet Evol. 2007;7:1–12. DOIPubMedGoogle Scholar
- Wormser GP, Liveris D, Nowakowski J, Nadelman RB, Cavaliere LF, McKenna D, Association of specific subtypes of Borrelia burgdorferi with hematogenous dissemination in early Lyme disease. J Infect Dis. 1999;180:720–5. DOIPubMedGoogle Scholar
- Jones KL, Glickstein LJ, Damle N, Sikand VK, McHugh G, Steere AC. Borrelia burgdorferi genetic markers and disseminated disease in patients with early Lyme disease. J Clin Microbiol. 2006;44:4407–13. DOIPubMedGoogle Scholar
- Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, Wang IN, Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect Immun. 1999;67:3518–24.PubMedGoogle Scholar
- Baranton G, Seinost G, Theodore G, Postic D, Dykhuizen D. Distinct levels of genetic diversity of Borrelia burgdorferi are associated with different aspects of pathogenicity. Res Microbiol. 2001;152:149–56. DOIPubMedGoogle Scholar
- Lagal V, Postic D, Ruzic-Sabljic E, Baranton G. Genetic diversity among Borrelia strains determined by single-strand conformation polymorphism analysis of the ospC gene and its association with invasiveness. J Clin Microbiol. 2003;41:5059–65. DOIPubMedGoogle Scholar
- Foretz M, Postic D, Baranton G. Phylogenetic analysis of Borrelia burgdorferi sensu stricto by arbitrarily primed PCR and pulsed-field gel electrophoresis. Int J Syst Bacteriol. 1997;47:11–8.PubMedGoogle Scholar
- Marti Ras N, Postic D, Baranton G. Borrelia burgdorferi sensu stricto, a bacterial species “Made in the U.S.A.”? Int J Syst Bacteriol. 1997;47:1112–7.PubMedGoogle Scholar
- Postic D, Ras NM, Lane RS, Humair P, Wittenbrink MM, Baranton G. Common ancestry of Borrelia burgdorferi sensu lato strains from North America and Europe. J Clin Microbiol. 1999;37:3010–2.PubMedGoogle Scholar
- Avise JC. Phylogeography: the history and formation of species. Cambridge (MA): Harvard University Press; 2000.
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–6. DOIPubMedGoogle Scholar
- Liveris D, Varde S, Iyer R, Koenig S, Bittker S, Cooper D, Genetic diversity of Borrelia burgdorferi in Lyme disease patients as determined by culture versus direct PCR with clinical specimens. J Clin Microbiol. 1999;37:565–9.PubMedGoogle Scholar
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. DOIPubMedGoogle Scholar
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. DOIPubMedGoogle Scholar
- Felsenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–6.
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–90. DOIPubMedGoogle Scholar
- Excoffier L, Laval G, Schneider S. Arlequin ver 3.0: an integratred software package for population data analysis. Evol Bioinform Online. 2005;1:47–50.
- Glockner G, Lehmann R, Romualdi A, Pradella S, Schulte-Spechtel U, Schilhabel M, Comparative analysis of the Borrelia garinii genome. Nucleic Acids Res. 2004;32:6038–46. DOIPubMedGoogle Scholar
- Sawyer S. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989;6:526–38.PubMedGoogle Scholar
- Wang G, van Dam AP, Dankert J. Evidence for frequent OspC gene transfer between Borrelia valaisiana sp. nov. and other Lyme disease spirochetes. FEMS Microbiol Lett. 1999;177:289–96. DOIPubMedGoogle Scholar
- Barbour AG, Fish D. The biological and social phenomenon of Lyme disease. Science. 1993;260:1610–6. DOIPubMedGoogle Scholar