Volume 21, Number 4—April 2015
Research
Sequence Variability and Geographic Distribution of Lassa Virus, Sierra Leone
Figure 1
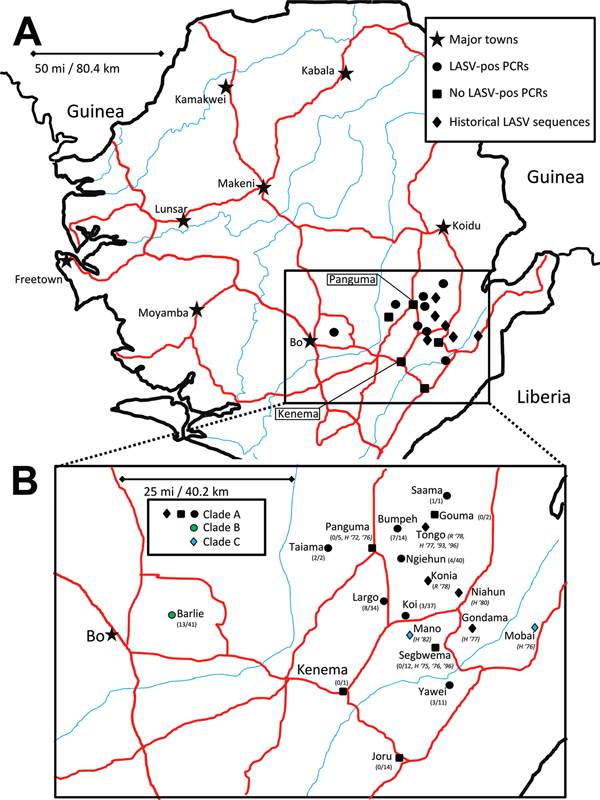
Figure 1. A) Locations of origin for Lassa virus (LASV) nucleic acid sequences, Sierra Leone. B) Enlarged view of region from which rodent specimens were collected. Major roads (red) and waterways (blue) are indicated. Symbols indicate major cities and towns (stars); sites in this study with rodent samples that were PCR positive for LASV (circles); sites in this study from which all samples from mulitmammate rats were PCR negative for LASV (squares); and sites from which published LASV sequences originated (diamonds). The color of the symbols in panel B indicates the clade for nucleoprotein sequence: black, clade A; green, clade B; blue, clade C. Fractions indicate, for each site included in this study, number of PCR-positive samples and total number of samples. Other designations for published sequence sites indicate type of isolate (H, human; R, rodent) and year(s) of isolation. No published information about geographic origin was available for the following strains: 807875, 331, 523, IJ531, Josiah, NL, SL06–2057, SL15, SL20, SL21, SL25, SL26, SL620.
1These authors contributed equally to this article.