Volume 21, Number 4—April 2015
Research
Sequence Variability and Geographic Distribution of Lassa Virus, Sierra Leone
Figure 2
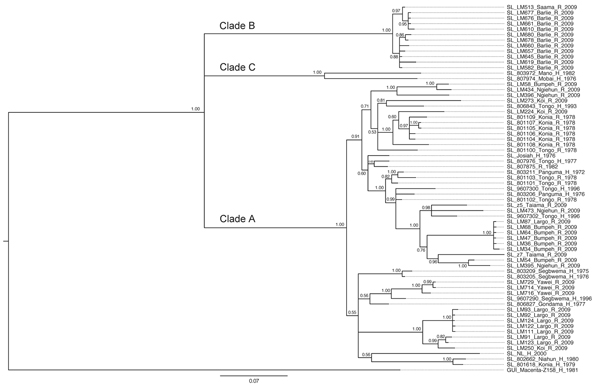
Figure 2. Phylogenetic analysis of Lassa virus isolates from Sierra Leone based on partial nucleoprotein (NP) gene sequences. The homologous NP fragments of 621 nt were aligned. The isolate Z-158, which originated from Macenta district in Guinea, was used as outgroup based on the previous phylogenetic analyses to root the tree. The 50% majority rule consensus tree was estimated by using Bayesian Inference method implemented in MrBayes software (32) using the Tamura 3-parameter substitution model with discrete γ-distributed rate variation. The strain labels contain information on the country of origin (SL, Sierra Leone; GUI, Guinea), strain designation, village or town of origin, type of isolate (H, human; R, rodent), and year of isolation. The numbers next to the branches indicate the posterior probability of particular clades. The clades as defined in this study (clades A, B, and C) are also indicated next to the appropriate branches. Scale bar indicates substitutions per site.
References
- Frame JD, Baldwin JM Jr, Gocke DJ, Troup JM. Lassa fever, a new virus disease of man from West Africa. I. Clinical description and pathological findings. Am J Trop Med Hyg. 1970;19:670–6 .PubMedGoogle Scholar
- Sogoba N, Feldmann H, Safronetz D. Lassa fever in West Africa: evidence for an expanded region of endemicity. Zoonoses Public Health. 2012;59(Suppl 2):43–7 . DOIPubMedGoogle Scholar
- Macher AM, Wolfe MS. Historical Lassa fever reports and 30-year clinical update. Emerg Infect Dis. 2006;12:835–7 . DOIPubMedGoogle Scholar
- Fichet-Calvet E, Rogers DJ. Risk maps of Lassa fever in West Africa. PLoS Negl Trop Dis. 2009;3:e388 . DOIPubMedGoogle Scholar
- Lukashevich IS, Clegg JC, Sidibe K. Lassa virus activity in Guinea: distribution of human antiviral antibody defined using enzyme-linked immunosorbent assay with recombinant antigen. J Med Virol. 1993;40:210–7 . DOIPubMedGoogle Scholar
- McCormick JB, Webb PA, Krebs JW, Johnson KM, Smith ES. A prospective study of the epidemiology and ecology of Lassa fever. J Infect Dis. 1987;155:437–44 . DOIPubMedGoogle Scholar
- McCormick JB. Lassa fever. In: Saluzzo JF, Dodet B, editors. Emergence and control of rodent-borne viral diseases. Amsterdam: Elsevier; 1999. p. 177–95.
- Richmond JK, Baglole DJ. Lassa fever: epidemiology, clinical features, and social consequences. BMJ. 2003;327:1271–5 . DOIPubMedGoogle Scholar
- Monath TP, Newhouse VF, Kemp GE, Setzer HW, Cacciapuoti A. Lassa virus isolation from Mastomys natalensis rodents during an epidemic in Sierra Leone. Science. 1974;185:263–5 . DOIPubMedGoogle Scholar
- Walker DH, Wulff H, Lange JV, Murphy FA. Comparative pathology of Lassa virus infection in monkeys, guinea-pigs, and Mastomys natalensis. Bull World Health Organ. 1975;52:523–34 .PubMedGoogle Scholar
- Smit A, van der Bank H, Falk T, de Castro A. Biochemical genetic markers to identify two morphologically similar South African Mastomys species (Rodentia: Muridae). Biochem Syst Ecol. 2001;29:21–30 . DOIPubMedGoogle Scholar
- Rosevear DR. Muridae: typical rats & mice, wood mice, fat mice, swamp rats. In: Rosevear DR, editor. Rodents of West Africa. London: Trustees of the British Museum (Natural History); 1969. p. 227–496.
- Peters CJ, Jahrling PB, Liu CT, Kenyon RH, McKee KT Jr, Barrera Oro JG. Experimental studies of arenaviral hemorrhagic fevers. Curr Top Microbiol Immunol. 1987;134:5–68 . DOIPubMedGoogle Scholar
- Ter Meulen J, Lukashevich I, Sidibe K, Inapogui A, Marx M, Dorlemann A, Hunting of peridomestic rodents and consumption of their meat as possible risk factors for rodent-to-human transmission of Lassa virus in the Republic of Guinea. Am J Trop Med Hyg. 1996;55:661–6 .PubMedGoogle Scholar
- Bowen MD, Rollin PE, Ksiazek TG, Hustad HL, Bausch DG, Demby AH, Genetic diversity among Lassa virus strains. J Virol. 2000;74:6992–7004 . DOIPubMedGoogle Scholar
- Safronetz D, Sogoba N, Lopez JE, Maiga O, Dahlstrom E, Zivcec M, Geographic distribution and genetic characterization of Lassa virus in sub-Saharan Mali. PLoS Negl Trop Dis. 2013;7:e2582 . DOIPubMedGoogle Scholar
- Ehichioya DU, Hass M, Becker-Ziaja B, Ehimuan J, Asogun DA, Fichet-Calvet E, Current molecular epidemiology of Lassa virus in Nigeria. J Clin Microbiol. 2011;49:1157–61 . DOIPubMedGoogle Scholar
- Lecompte E, Fichet-Calvet E, Daffis S, Koulemou K, Sylla O, Kourouma F, Mastomys natalensis and Lassa fever, West Africa. Emerg Infect Dis. 2006;12:1971–4 . DOIPubMedGoogle Scholar
- Günther S, Emmerich P, Laue T, Kuhle O, Asper M, Jung A, Imported Lassa fever in Germany: molecular characterization of a new Lassa virus strain. Emerg Infect Dis. 2000;6:466–76 . DOIPubMedGoogle Scholar
- Günther S, Weisner B, Roth A, Grewing T, Asper M, Drosten C, Lassa fever encephalopathy: Lassa virus in cerebrospinal fluid but not in serum. J Infect Dis. 2001;184:345–9 . DOIPubMedGoogle Scholar
- Trappier SG, Conaty AL, Farrar BB, Auperin DD, McCormick JB, Fisher-Hoch SP. Evaluation of the polymerase chain reaction for diagnosis of Lassa virus infection. Am J Trop Med Hyg. 1993;49:214–21 .PubMedGoogle Scholar
- Olschläger S, Lelke M, Emmerich P, Panning M, Drosten C, Hass M, Improved detection of Lassa virus by reverse transcription–PCR targeting the 5′ region of S RNA. J Clin Microbiol. 2010;48:2009–13 . DOIPubMedGoogle Scholar
- Demby AH, Chamberlain J, Brown DW, Clegg CS. Early diagnosis of Lassa fever by reverse transcription–PCR. J Clin Microbiol. 1994;32:2898–903 .PubMedGoogle Scholar
- Mills JN, Childs JE, Ksiazek TG, Peters CJ, Wallis MV. Methods for trapping and sampling small mammals for virologic testing. Atlanta: US Department of Health and Human Services; 1995.
- Lecompte E, Brouat C, Duplantier J-M, Galan M, Granjon L, Loiseau A, Molecular identification of four cryptic species of Mastomys (Rodentia, Murinae). Biochem Syst Ecol. 2005;33:681–9. DOIGoogle Scholar
- Vieth S, Drosten C, Lenz O, Vincent M, Omilabu S, Hass M, RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg. 2007;101:1253–64 . DOIPubMedGoogle Scholar
- Leski TA, Lin B, Malanoski AP, Wang Z, Long NC, Meador CE, Testing and validation of high density resequencing microarray for broad range biothreat agents detection. PLoS ONE. 2009;4:e6569 . DOIPubMedGoogle Scholar
- Metzgar D, Myers CA, Russell KL, Faix D, Blair PJ, Brown J, Single assay for simultaneous detection and differential identification of human and avian influenza virus types, subtypes, and emergent variants. PLoS ONE. 2010;5:e8995 . DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9 . DOIPubMedGoogle Scholar
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5 . DOIPubMedGoogle Scholar
- Vieth S, Torda AE, Asper M, Schmitz H, Gunther S. Sequence analysis of L RNA of Lassa virus. Virology. 2004;318:153–68 . DOIPubMedGoogle Scholar
- Lalis A, Leblois R, Lecompte E, Denys C, Ter Meulen J, Wirth T. The impact of human conflict on the genetics of Mastomys natalensis and Lassa virus in West Africa. PLoS ONE. 2012;7:e37068 . DOIPubMedGoogle Scholar
- ter Meulen J, Lenz O, Koivogui L, Magassouba N, Kaushik SK, Lewis R, Short communication: Lassa fever in Sierra Leone: UN peacekeepers are at risk. Trop Med Int Health. 2001;6:83–4 . DOIPubMedGoogle Scholar
- Shaffer JG, Grant DS, Schieffelin JS, Boisen ML, Goba A, Hartnett JN, Lassa fever in post-conflict Sierra Leone. PLoS Negl Trop Dis. 2014;8:e2748 . DOIPubMedGoogle Scholar
1These authors contributed equally to this article.