Volume 21, Number 4—April 2015
Dispatch
Pathogenicity of 2 Porcine Deltacoronavirus Strains in Gnotobiotic Pigs
Abstract
To verify whether porcine deltacoronavirus infection induces disease, we inoculated gnotobiotic pigs with 2 virus strains (OH-FD22 and OH-FD100) identified by 2 specific reverse transcription PCRs. At 21–120 h postinoculation, pigs exhibited severe diarrhea, vomiting, fecal shedding of virus, and severe atrophic enteritis. These findings confirm that these 2 strains are enteropathogenic in pigs.
Porcine epidemic diarrhea virus (PEDV) (family Coronaviridae, genus Alphacoronavirus) was discovered in the United States in May 2013. The virus has now spread nationwide and caused a high number of deaths among suckling pigs (1,2). In regions of the United States to which PEDV is epidemic, a new coronavirus, genetically distinct from PEDV, porcine deltacoronavirus (PDCoV) (genus Deltacoronavirus), has been simultaneously and frequently detected in diarrheic fecal samples from pigs (3–5).
The clinical role and disease severity of PDCoV in the field is reportedly less than that of PEDV (6). To confirm the role of PDCoV as an enteric viral pathogen and understand disease progression, we studied the pathogenicity of 2 strains of PDCoV (OH-FD22 and OH-FD100) in gnotobiotic pigs. We developed in situ hybridization and immunofluorescence staining methods to verify the tissue sites of PDCoV replication in infected pigs.
In February and July 2014, intestinal contents were obtained from young nursing piglets with diarrhea on farms in Ohio, USA. PDCoV strains OH-FD22 and OH-FD100 were detected in samples by using a TaqMan quantitative reverse transcription RT-PCR (qRT-PCR) specific for the membrane gene (nt 23395–23466) as reported (7), or an RT-PCR specific for the PDCoV membrane gene (nt 23111–23651) based on the sequence of PDCoV strain (USA/Illinois121/2014; GenBank accession no. KJ481931), as described in the in situ hybridization method. The partial membrane gene sequences of OH-FD22 and OH-FD100 were identical to that of USA/Illinois121/2014. Samples were negative for PEDV, rotavirus groups A–C, transmissible gastroenteritis virus (TGEV)/porcine respiratory coronavirus (PRCV), and caliciviruses (noroviruses, sapoviruses, and St. Valerien–like viruses) by RT-PCR as reported (8). The samples were bacteriologically sterilized by using 0.22-μm syringe filters and then prepared as inoculum.
Near-term gnotobiotic pigs were delivered aseptically by hysterectomy from 2 specific pathogen–free sows (9). Seven 11- to 14-day-old pigs were randomly assigned to a PDCoV-inoculated group (pigs 1–5) or as negative controls (pigs 6 and 7). Pigs 1–3 and pigs 4 and 5 were inoculated orally with 8.8 log10 genomic equivalents (GEs) of PDCoV strain OH-FD22 and 11.0 log10 GEs of OH-FD100, respectively. Clinical signs were monitored hourly. Pig 2 was monitored for long-term clinical signs and virus shedding until day postinoculation (dpi) 23. Pigs were euthanized for pathologic examination at 24–48 h or >48 h after onset of clinical signs. All animal-related experimental protocols were approved by the Ohio State University Institutional Animal Care and Use Committee.
Fecal or rectal swab samples were prepared as described (8,9). Virus RNA was extracted by using the Mag-MAX Viral RNA Isolation Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Titers of virus shed in feces were determined by using the qRT-PCR and the OneStep RT-PCR Kit (QIAGEN, Valencia, CA, USA) as reported (7). A standard curve was generated by using the PCR amplicon (nt 23111–23651) of strain OH-FD22. The detection limit of the qRT-PCR was 10 GEs/reaction, which corresponded to 4.6 log10 and 3.6 log10 GEs/mL of PDCoV in fecal and serum samples, respectively.
Small and large intestinal tissues and other major organs (lung, liver, heart, kidney, spleen, and mesenteric lymph nodes) were examined. Mean jejunal ratios of villous height to crypt depth were measured as reported (8). For PDCoV RNA detection in formalin-fixed, paraffin-embedded tissues, a nonradioactive digoxigenin-labeled cDNA probe specific for the 541-bp virus membrane gene sequence (nt 23111–23651), amplified with the primers forward 5′-CGCGTAATCGTGTGATCTATGT-3′ and reverse 5′-CCGGCCTTTGAAGTGGTTAT-3′, was used for in situ hybridization as described (10). Reverse transcription was conducted at 50°C for 30 min, followed by denaturation at 94°C for 5 min; 35 cycles at 94°C for 40 s, 55°C for 40 s, and 72°C for 1 min; and final extension at 72°C for 7 min.
OH-FD22–infected pig 2 was immunized intramuscularly with OH-FD22 from the gnotobiotic pig–passaged intestinal contents that were semipurifed by sucrose gradient ultracentrifugation (11) and mixed with complete and incomplete Freund’s adjuvants at dpi 30 and dpi 44 (11). Immunofluorescence staining was performed on frozen or formalin-fixed, paraffin-embedded tissues as described (8,9) by using hyperimmune gnotobiotic pig antiserum against OH-FD22. Tissues from control pigs 6 and 7 and PEDV-infected gnotobiotic pigs (8) were used negative controls for in situ hybridization/immunofluorescence staining.
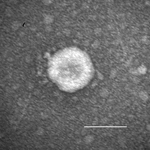
Acute, severe, watery diarrhea, vomiting, or both developed in all inoculated pigs. Clinical signs developed at hour postinoculation (hpi) 21–24, regardless of the inoculum strain or dose (Table 1). At hpi 96–120, pig 1 exhibited severe dehydration, loss of bodyweight, and lethargy. Pig 2, which was followed up longer, showed diarrhea until dpi 7. All inoculated pigs exhibited onset of clinical disease similar to that of infection with PEDV strain PC21A (6.3–9.0 log10 GEs/pig) in gnotobiotic pigs (8). Immune electron microscopy with hyperimmune serum to PDCoV from a gnotobiotic pig showed only PDCoV particles in the intestinal contents (Figure 1). For pig-passaged OH-FD22 and OH-FD100 samples, RT-PCR results were negative for PEDV, rotavirus groups A–C, TGEV/PRCV, and caliciviruses. Detection of fecal virus shedding at hpi 24 coincided with onset of clinical signs (Table 1) in pigs 2–5. In pig 1, which showed only vomiting at hpi 24, fecal shedding occurred at hpi 48 at the onset of diarrhea.
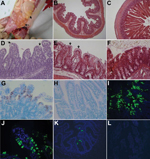
Macroscopic examination showed that all infected pigs had PEDV-like lesions characterized by thin and transparent intestinal walls (proximal jejunum to colon) and accumulation of large amounts of yellow fluid in the intestinal lumen (Figure 2, panel A). The stomach was filled with curdled milk. Other internal organs appeared normal. Histologic lesions included acute diffuse, severe atrophic enteritis (Figure 2, panels B, D) and mild vacuolation of superficial epithelial cells in cecum and colon (Figure 2, panel E). The mean jejunal ratios of villous height to crypt depth of infected pigs 3–5 at hpi 72–120 ranged from 1.4 to 3.6 (Table 2), which were similar to those in gnotobiotic pigs experimentally infected with PEDV strain PC21A (8). Clinical signs or lesions did not develop in negative control pigs during the experiment (Figure 2, panels C, F).
In situ hybridization–positive or immunofluorescence-stained cells were observed mainly in the villous epithelium of small (duodenum to ileum) and large intestines (Table 2; Figure 2, panels G, I–K). Immunofluorescence was confined to the cytoplasm of villous epithelial cells (Figure 2, panel J) and was infrequently observed in crypt epithelial cells. No other internal organs of infected pigs showed in situ hybridization–positive or immunofluorescence-positive staining. In situ hybridization–positive or immunofluorescence-stained cells were not detected in negative control pigs (Figure 2, panels H, L) and PEDV-infected gnotobiotic pigs.
Under the experimental conditions used, no PDCoV-inoculated pigs at hpi 72–168 had detectable virus RNA (<3.6 log10 GEs/mL) in serum. However, viremia was detected frequently in symptomatic PEDV-infected pigs (8,13).
Since 2013–2014, newly emerged PEDV and PDCoV have spread throughout the United States and caused a high number of pig deaths (1,2,6), but no studies of the pathogenicity of PDCoV have been reported. Our data confirm that PDCoV strains OH-FD22 and OH-FD100 are enteropathogenic and acutely infect the entire intestine. However, the jejunum and ileum are the primary sites of infection. PDCoV infection caused severe atrophic enteritis accompanied by severe diarrhea, vomiting, or both. These clinical and pathologic features in PDCoV-infected pigs resemble those of PEDV and TGEV infections. Differential diagnosis of PDCoV, PEDV, and TGEV is critical to control virus epidemic diarrhea on swine farms in the United States.
Acknowledgments
We thank J. Hanson, R. Wood, and J. Ogg for assisting with animal care; Q. Wang for providing advice on qRT-PCR for PDCoV; and X. Wang and M. Lee for providing technical assistance.
Salaries and research support were provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, Ohio State University. This study was supported by Four Star Animal Health Inc. (Loramie, OH, USA) (L.J.S.) and Ohio Agricultural Research and Development Center SEEDS Program (grant OAOH1536 to K.J.).
Dr. Jung is a veterinary pathologist at Ohio State University, Wooster, Ohio. His research interests include diagnostic molecular pathology and pathogenesis of virus infections and analysis of immune responses to swine enteric viral infections by using germ-free or conventional pig models.
References
- Cima G. Viral disease affects U.S. pigs: porcine epidemic diarrhea found in at least 11 states. J Am Vet Med Assoc. 2013;243:30–1 .PubMedGoogle Scholar
- Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. 2013;25:649–54. DOIPubMedGoogle Scholar
- Marthaler D, Jiang Y, Collins J, Rossow K. Complete genome sequence of strain SDCV/USA/Illinois121/2014, a porcine deltacoronavirus from the United States. Genome Announc. 2014;2:e00218–14.
- Wang L, Byrum B, Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis. 2014;20:1227–30. DOIPubMedGoogle Scholar
- Li G, Chen Q, Harmon KM, Yoon KJ, Schwartz KJ, Hoogland MJ, Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc. 2014;2:e00278–14.
- Marthaler D, Raymond L, Jiang Y, Collins J, Rossow K, Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg Infect Dis. 2014;20:1347–50. DOIPubMedGoogle Scholar
- Jung K, Wang Q, Scheuer KA, Lu Z, Zhang Y, Saif LJ. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg Infect Dis. 2014;20:662–5. DOIPubMedGoogle Scholar
- Jung K, Wang Q, Kim Y, Scheuer K, Zhang Z, Shen Q, The effects of simvastatin or interferon-alpha on infectivity of human norovirus using a gnotobiotic pig model for the study of antivirals. PLoS ONE. 2012;7:e41619. DOIPubMedGoogle Scholar
- Jung K, Ha Y, Chae C. Pathogenesis of swine influenza virus subtype H1N2 infection in pigs. J Comp Pathol. 2005;132:179–84. DOIPubMedGoogle Scholar
- Guo M, Qian Y, Chang KO, Saif LJ. Expression and self-assembly in baculovirus of porcine enteric calicivirus capsids into virus-like particles and their use in an enzyme-linked immunosorbent assay for antibody detection in swine. J Clin Microbiol. 2001;39:1487–93. DOIPubMedGoogle Scholar
- Coussement W, Ducatelle R, Debouck P, Hoorens J. Pathology of experimental CV777 coronavirus enteritis in piglets. I. Histological and histochemical study. Vet Pathol. 1982;19:46–56. DOIPubMedGoogle Scholar
- Opriessnig T, Xiao CT, Gerber PF, Zhang J, Halbur PG. Porcine epidemic diarrhea virus RNA present in commercial spray-dried porcine plasma is not infectious to naive pigs. PLoS ONE. 2014;9:e104766 . DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 21, Number 4—April 2015
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Kwonil Jung or Linda J. Saif, Food Animal Health Research Program, Ohio Agricultural Research and Development Center, Department of Veterinary Preventive Medicine, Ohio State University, 1680 Madison Ave, Wooster, OH 44691, USA; or
Top