Volume 22, Number 12—December 2016
Research
African Horse Sickness Caused by Genome Reassortment and Reversion to Virulence of Live, Attenuated Vaccine Viruses, South Africa, 2004–2014
Figure 4
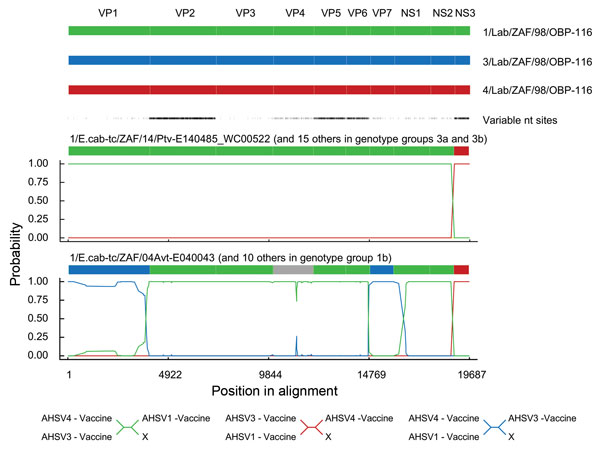
Figure 4. Statistical evidence of reassortment within the genomes of African horse sickness (AHS) virus field isolates identified in outbreaks in the AHS controlled area in Western Cape Province, South Africa, 2004–2014. A hidden Markov model–based approach (BURT-HMM) was used to classify individual nucleotides within each of the 10 segments of individual AHS virus isolates into 3 different categories: 1/Lab/ZAF/98/OBP-116-like (green), 3/Lab/ZAF/98/OBP-116-like (blue), and 4/Lab/ZAF/98/OBP-116-like (red). Probability supports for these classifications yielded by the BURT-HMM with the highest likelihood are plotted along the genome. Positions of segment boundaries are given in the diagram above the plots. The phylogenetic clusterings that are implied by differently colored segments in these plots are indicated below the plots. The segment indicated in gray could not be convincingly classified because it closely resembles both 1/Lab/ZAF/98/OBP-116 and 3/Lab/ZAF/98/OBP-116.