Spread of Canine Influenza A(H3N2) Virus, United States
Ian E.H. Voorhees, Amy L. Glaser, Kathy L. Toohey-Kurth, Sandra Newbury, Benjamin D. Dalziel, Edward Dubovi, Keith Poulsen, Christian Leutenegger, Katriina J.E. Willgert, Laura Brisbane-Cohen, Jill Richardson-Lopez, Edward C. Holmes, and Colin R. Parrish

Author affiliations: Cornell University, Ithaca, New York, USA (I.E.H. Voorhees, A.L. Glaser, E.J. Dubovi, K.J.E. Willgert, L. Brisbane-Cohen, C.R. Parrish); University of Wisconsin, Madison, Wisconsin, USA (K. Toohey-Kurth, S. Newbury, K. Poulsen); Oregon State University, Corvallis, Oregon, USA (B.D. Dalziel); IDEXX Laboratories, West Sacramento, California, USA (C. Leutenegger); Royal Veterinary College, London, UK (K.J.E. Willgert); Merck Animal Health, Madison, New Jersey, USA (J. Richardson-Lopez); University of Sydney, Sydney, New South Wales, Australia (E.C. Holmes)
Main Article
Figure 4
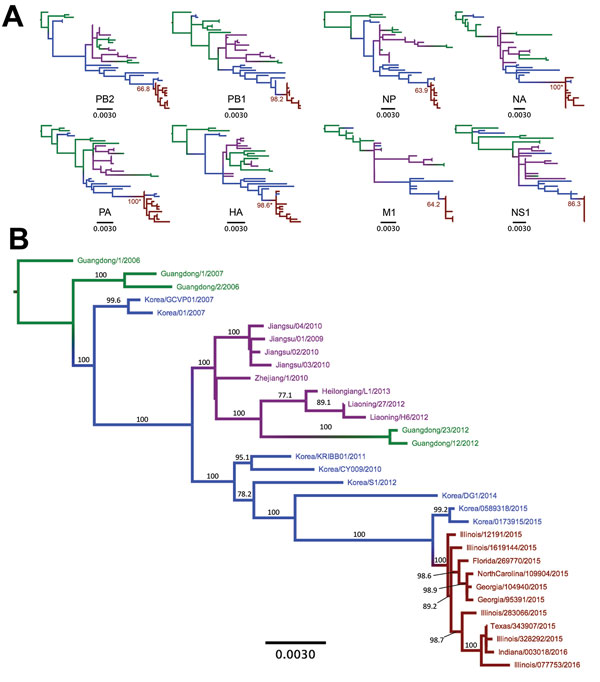
Figure 4. Phylogenetic trees of canine influenza A(H3N2) virus (H3N2 CIV) sequences showing the initial emergence of the virus in southern China (green branches), its appearance in northern and eastern China (magenta branches) and South Korea (blue branches), and its introduction into the United States (red branches). A) Individual genome segment sequences. Red branch numbers indicate bootstrap proportion of US H3N2 CIV clade. Asterisks indicate polyphyletic clades containing US strains and most recent strains from South Korea. B) Concatenated segment phylogenies of all available complete nonreassortant H3N2 CIV genomes. Branch number indicates bootstrap proportions >75. All branch lengths are proportional to the number of nucleotide substitutions per site. All trees rooted by using sequences from the earliest isolated H3N2 CIV. Scale bars indicate nucleotide substitutions per site. HA, hemagglutinin; M1, matrix 1; NA, neuraminidase; NP, nucleocapsid protein; NS1, nonstructural 1; PA, polymerase acidic; PB1, polymerase basic 1; PB2, polymerase basic 2.
Main Article
Page created: November 16, 2017
Page updated: November 16, 2017
Page reviewed: November 16, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
