Volume 24, Number 8—August 2018
Research
Human Norovirus Replication in Human Intestinal Enteroids as Model to Evaluate Virus Inactivation
Figure 4
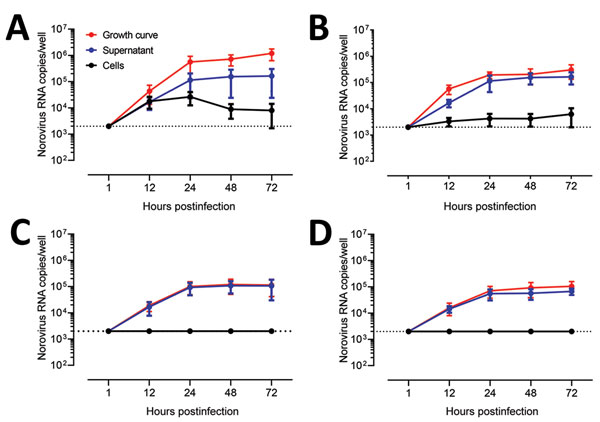
Figure 4. Evaluation as human norovirus replication in human intestinal enteroids (HIEs) by assessment of kinetics of infection for 4 GII genotypes. We inoculated jejunal HIE monolayers (J3 line) with A) GII.4 Pe-GII.4 Sydney (3.3 × 105 RNA copies/well), B) GII.P12-GII.3 (5.3 × 105 RNA copies/well), C) GII.P16-GII.2 (3.2 × 105 RNA copies/well), or D) GII.Pg-GII.1 (9.3 × 105 RNA copies/well). After 1 h at 37°C in 5% CO2, monolayers were washed, and media was replaced with differentiation media and incubated for 3 d. For the growth curve, we extracted RNA from frozen lysates (cells and supernatant) at the indicated time points. For the cells vs. supernatant experiment, we removed supernatants by centrifugation before harvesting the cells. Data represent mean ± SD of 2 experiments with 3 wells for each time point. Dotted lines represent RT-quantitative PCR limit of detection.