Volume 26, Number 1—January 2020
Synopsis
Candidatus Mycoplasma haemohominis in Human, Japan
Figure 5
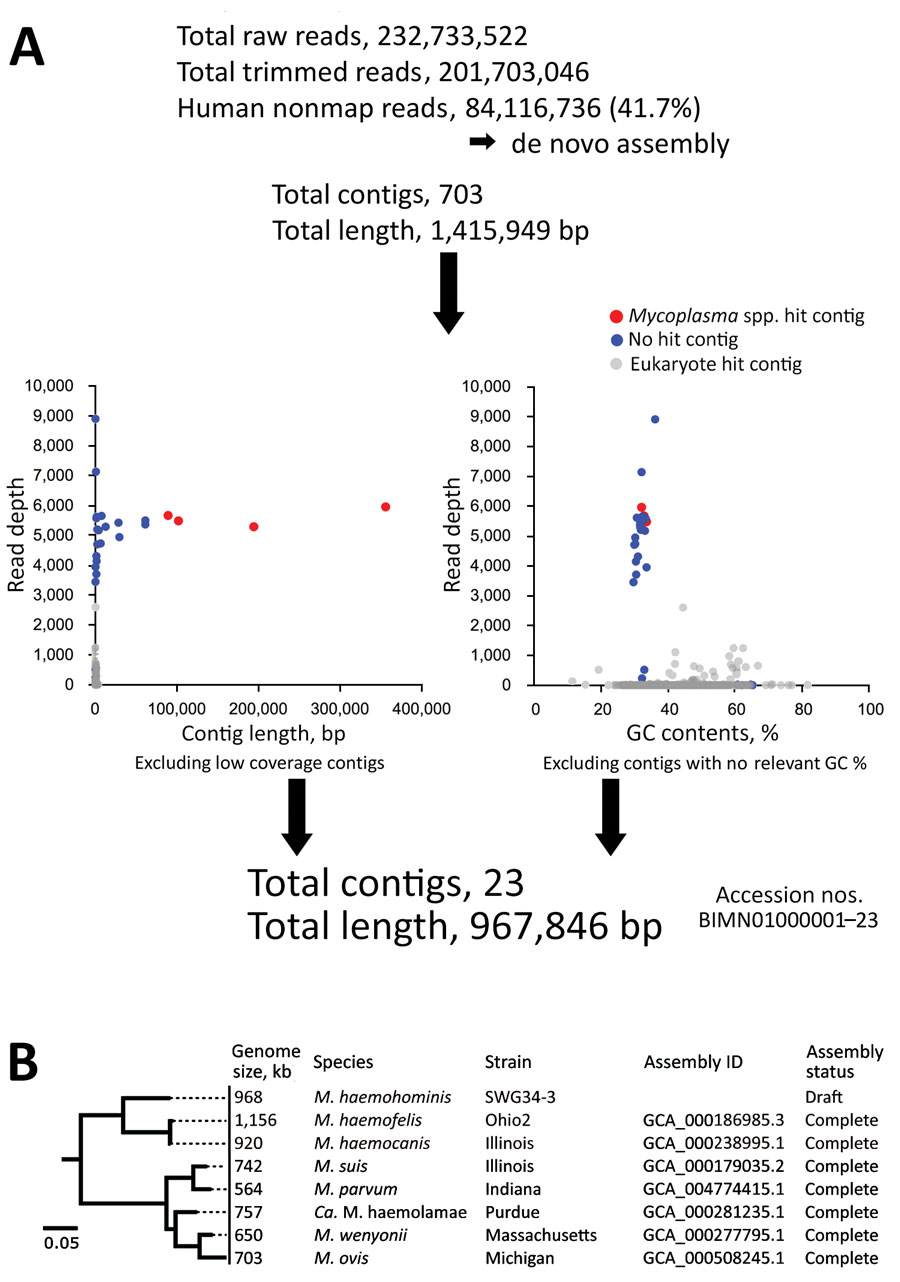
Figure 5. Analysis for Candidatus Mycoplasma haemohominis in serum of a 42-year-old man, Japan. A) Prediction that de novo assemblies contained bacteria and human DNA sequences. Bacteria-related sequences were identified by using read depth, % GC, and blastn (https://blast.ncbi.nlm.nih.gov) search results. Read depth indicates how many times next-generation sequencing confirmed the sequence at each nucleotide position. B) Phylogenetic tree of 16S rRNA genes of Mycoplasma spp. The tree was constructed by using FastTree version 2.1.10 (http://www.microbesonline.org). Scale bar indicates nucleotide substitutions per site. ID, identification.
1These authors contributed equally to this article.
Page created: December 18, 2019
Page updated: December 18, 2019
Page reviewed: December 18, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.