Volume 26, Number 1—January 2020
Synopsis
Candidatus Mycoplasma haemohominis in Human, Japan
Cite This Article
Citation for Media
Abstract
Hemotropic mycoplasmas are common pathogens in animals, but it remains unclear what role these pathogens play in human infections. We report clinical and biologic characterization of Candidatus Mycoplasma haemohominis infection in a 42-year-old man in Japan. The patient had severe hemophagocytic syndrome 1 month after an accidental needlestick injury. Metagenomic deep sequencing identified Candidatus M. haemohominis and determined its draft genome for an isolate from serum of the patient. A high copy number of the Candidatus M. haemohominis genome was detected in serum and bone marrow samples. Electron microscopy examination showed morphologic characteristics of Candidatus M. haemohominis. Levofloxacin monotherapy induced resistance caused by a gyrase A gene mutation in the quinolone resistance–determining region, but a combination treatment with moxifloxacin and minocycline was effective. We identified Candidatus M. haemohominis in a patient who had life-threatening symptoms related to multiple organ infection. Human infection with this mycoplasma might occur more frequently than has been generally recognized.
Hemotropic mycoplasmas (or hemoplasmas; size <1 μm) are unculturable, cell wall–deficient, gram-negative bacteria that parasitize on the surface of the erythrocytes of numerous domestic and wild animals, such as cats, dogs, rodents, swine, cattle, sheep, bears, and bats (1–4). These pathogens can cause pyrexia, hemolytic anemia, and icterus (1,5). There have been few studies on molecular characterization of hemoplasmas to confirm infections in humans (6–11), possibly because hemoplasmas are unculturable and are liable to be overlooked by physicians (11,12). Steer et al. found that the putative species Mycoplasma species Candidatus Mycoplasma haemohominis can infect humans and cause hemolytic anemia and pyrexia (9). However, only a partial sequence of Candidatus M. haemohominis (GenBank accession no. GU562825) could be confirmed (9). Moreover, clinical manifestations of Candidatus M. haemohominis infections in humans have not been well characterized.
We identified and characterized Candidatus M. haemohominis infections in a patient with pyrexia of unknown origin. The patient had various life-threatening symptoms that were not limited to hemolytic anemia and was infected with this bacterium after an accidental needlestick injury. We also analyzed the genome of Candidatus M. haemohominis isolated from specimens obtained from the patient. This study was approved by the Institutional Review Board of Showa University (Tokyo, Japan) and the National Institute of Infectious Diseases (Tokyo). We obtained informed consent from the patient.
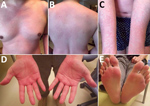
The case-patient was a 42-year-old man (physician) who had no unusual medical history and no recent overseas travel history. The patient was admitted to Showa University School of Medicine Hospital (Tokyo, Japan) because of pyrexia, anemia, and liver dysfunction. One month before admission, he had accidentally pricked his finger when performing needle biopsy of the liver for 1 inpatient, who was admitted to this hospital because of cryptogenic liver injury and anemia after traveling overseas. Two weeks after the needlestick injury, the case-patient had pyrexia and whole-body erythema with pruritus (Figure 1). His rash disappeared after 3 days. However, he was admitted to the hospital because lymphadenopathy, hepatosplenomegaly, and pyrexia developed.
We provide the clinical course for the case-patient (Figure 2). At admission, the case-patient had relative bradycardia (96 beats/min) and a body temperature of 39.5°C. Laboratory results showed an increase in levels of aspartate aminotransferase (274 U/L, reference range 10–40 U/L), lactate dehydrogenase (664 U/L, reference range 120–245 U/L), ferritin (8,748 ng/mL, reference range 20–400 ng/mL), soluble interleukin-2 receptor (8,791 U/mL, reference range 122–496 U/mL), and C-reactive protein (8.45 mg/dL, reference range 0.00–0.20 mg/dL). A complete blood count showed anemia (hemoglobin concentration 11.9 g/dL, reference range 13.6–18.3 g/L), but the leukocyte count (4.8 × 103 cells/μL, reference range 3.5–9.0 × 103 cells/μL) and platelet count (16.5 × 104/μL, reference range 14.0–37.9 × 104 cells/μL) were within reference intervals. A Coombs test result was negative, but low haptoglobin concentrations (<8 mg/dL, range 30–200 mg/dL) were found.
Test results were negative for hepatitis A, B, C, and E viruses; measles virus; rubella virus; parvovirus; and HIV. Results of antinuclear antibody (titer 1:80) and smooth muscle antibody (titer 1:40) tests were positive, but test results for antimitochondrial M2 antibody, mitochondrial antibody, double-stranded DNA antibody, and lupus anticoagulant were negative. Epstein-Barr virus (EBV) DNA load determined by PCR was 3.0 × 102 copies/mL. However, Southern blot hybridization did not detect clonality of EBV-infected cells. Levels of herpes simplex virus, human herpesvirus 6 and 8, varicella zoster virus, and cytomegalovirus were below reference values, as determined by PCR. The serum IgG level was 3,967 mg/dL (reference range 800–1,750 mg/dL). However, the result of a serum-free light chain test was within the reference limit.
Our initial diagnosis was hemophagocytic syndrome (HPS) related to undetermined disease, and the patient was given steroid pulse therapy (1,000 mg) on day 1. Because his symptoms persisted, he was given etoposide (100 mg/m2), cyclosporine (5 mg/kg), and dexamethasone (20 mg) on day 7. After this treatment was started, whole-genome sequencing of a peripheral blood sample detected a novel hemotropic Mycoplasma sp.
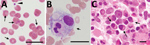
On day 11, we found a prolonged activated partial thromboplastin time (50.6 s, reference range 25–45 s), normal prothrombin time (12.6 s), low coagulation factor VIII activity (1.8%, reference range 78%–165%), and low von Willebrand factor (VWF) activity (ristocetin cofactor <10%, reference range 50%–150%). Moreover, serum levels of fibrinogen, antithrombin III, and disintegrin and metalloproteinase with thrombospondin type 1 motif, member 13 were within reference ranges. Microscopic examination of a Giemsa-stained blood smear showed coccoid forms on the erythrocyte surface (Figure 3, panel A). A bone marrow aspirate showed hemophagocytosis and increased levels of reactive plasma cells (19.5%) (Figure 3, panels B, C). On day 12, hypoglycemia (glucose level <5 mg/dL, reference range, 61–139 mg/dL) was noted in blood from the collection tube used for serologic tests, but the glucose level was within the reference range for blood in a container that contained citric acid with NaF.
On day 13, the case-patient received a diagnosis of Candidatus M. haemohominis infection–associated HPS and was given levofloxacin. Hypoglycemia improved after administration of levofloxacin. However, pyrexia and anemia developed, and coccoid forms on erythrocytes in peripheral blood and bone marrow were again observed on day 37. On day 46, he was given moxifloxacin and minocycline because of suspected bacterial resistance to levofloxacin. After this treatment was initiated, his symptoms, such as pyrexia, anemia, and hypoglycemia, promptly resolved, and the patient was discharged on day 61. One year after discontinuation of treatment, the patient remained well, and no Candidatus M. haemohominis DNA was detected in his serum.
High-Throughput Unbiased RNA Sequencing
We purified total RNA from a patient serum sample by using the RNeasy Mini Kit (QIAGEN, https://www.qiagen.com). We prepared RNA-Seq libraries by using the ScriptSeq version 2 RNA-Seq Library Preparation Kit (Illumina, https://www.illumina.com) and sequenced these libraries as single-end 151-mers by using the NextSeq 500 sequencer (Illumina) (13,14).
Whole-Genome Analysis
We performed metagenomic short-read DNA sequencing by using NextSeq 500 (Illumina) with DNA extracted from serum specimens. We excluded human-related short reads by using a Burrows–Wheeler Mapping Program with default parameters against human genome sequences (GRCh38.p13) (15).
We obtained a draft genome sequence of Candidatus M. haemohominis by de novo assembly using metagenomic DNA-Seq short reads. First, we excluded human genomic DNA sequences (≈58.3%) by using read-mapping analysis, followed by de novo assembly with the remaining short reads. The total number of contigs was 703, and total length was ≈1.42 Mb, suggesting that whole contigs included bacterial and human-related DNA sequences. Therefore, to extract the bacterial sequences, we considered read depth and coverage, % GC of each contig, and a blastn search (https://blast.ncbi.nlm.nih.gov). A total of 23 contigs showed a marked read depth coverage (average × 5,500) and 30% GC content, although blastn search analysis showed that 4 contigs were assigned to the Mycoplasma genome and 19 contigs were assigned to unknown sequences.
Electron Microscopy
We used negative staining for serum samples. Small aliquots of serum samples were absorbed onto glow-discharged, 300-mesh, heavy-duty carbon-coated copper Cu grids (Veco Grids; Nisshin EM, http://nisshin-em.co.jp) for 2 min, and excess liquid was blotted with Whatman filter paper (GE Healthcare, https:///www.gehealthcare.com). We then washed grids twice with Milli-Q water (http://emdmillipore.com) and negatively stained them with 2% uranyl acetate. We observed specimens by using an H7700 transmission electron microscope (Hitachi, https://www.hitachi.com) at 80 kV and ×10,000 magnification.
We fixed tissue samples with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 mol/L phosphate buffer (pH 7.4) for 2 h at room temperature, postfixed these samples in 1% osmium tetroxide, and embedded them in Epon resin. We stained ultrathin sections with uranyl acetate and lead citrate and observed them under a transmission electron microscope (HT7700; Hitachi) at 80 kV.
In Situ Hybridization
We used 16S and 23S rRNA genes for in situ hybridization analysis. We amplified a target fragment by using PCR with digoxigenin-11-dUTP and MHaemohominis1F (5′-AATTAACGCTGATGGCATGC-3′) and MHaemohominis600R (5′- TCCTACCGTATTCTAGACGGAC-3′) primers. We purified a PCR amplicon by using a PCR Purification Kit (QIAGEN) and denatured it by heat shock before hybridization. We treated deparaffinized slides with 0.3% H2O2/methanol for 30 min and 0.2 mol/L HCl for 20 min, then incubated slides with proteinase K (3–10 μg/mL) for 30 min at 37°C. After prehybridization, we hybridized slides with 2 pmol/L of denatured probe per slide in hybridization buffer (20% formamide; 5× saline–sodium citrate [SSC], 5 × Denhardt solution; 50 mmol/L HEPES buffer [pH 7.0], and 40 µg/mL salmon sperm DNA) overnight at 42°C. We washed the slides twice with 2× SSC at 50°C for 15 min and twice with 0.2× SSC at 50°C for 15 min, added anti-digoxigenin monoclonal antibody (Sigma, https://www.sigmaaldrich.com), and incubated the slides for 45 min. We than amplified signals by using a GenPoint Kit (Dako Agilent, https://www.agilent.com) and detected these signals by using 3,3′-diaminobenzidine as a chromogen.
Real-Time PCR
We used a DNA fragment of the 16S rRNA gene of Candidatus M. haemohominis in a Taqman real-time PCR (16). We then performed PCR amplification in 25-μL reaction mixtures containing QuantiTect Probe PCR Master Mixture (QIAGEN), 0.4 μmol/L of each primer, 0.2 μmol/L of TaqMan probe, and 100 ng of isolated DNA. PCR conditions were 95°C for 15 min, followed by 45 cycles of 94°C for 15 s and 60°C for 1 min in a Mx3005P Apparatus (Dako Agilent).
Sequence Data
We deposited metagenomic short-read sequences for DNA-Seq in the DDBJ (BioProject PRJDB7871; BioSample SAMD00156464; DRR accession no. DRR164892). We deposited the draft, annotated genome sequence of Candidatus M. haemohominis SWG34-3 in the DNA Data Bank of Japan (accession nos. SAMD00156495 and BIMN01000001–23). The 16S rRNA sequence was deposited under accession no. MHSWG343_r0010. The predicted gyrase A gene nucleotide sequence was deposited under accession no. MHSWG343_02220, and the coding sequence was deposited under accession no. GCE63237.1.
Identification and Whole-Genome Sequencing of Candidatus M. haemohominis
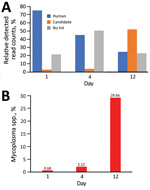
Metagenomic deep RNA sequence analysis strongly suggested that rather than other pathogens or virus infections, the Mycoplasma spp. could be associated with signs and symptoms of the case-patient. RNA-Seq short reads related to the Mycoplasma spp. were increasingly detected in the serum of the case-patient (Figure 4). A total of 41.7% of human-unmapped reads were extracted and then subjected to de novo assembly to identify the mycoplasma draft genome (23 contigs, 967,846 bp) (Figure 5, panel A). The draft genome sequence identified a potential pathogen that could be Candidatus M. haemohominis on the basis of 16S rRNA sequence homology. Also, this pathogen showed similarity with closely related species, such as M. haemofelis and M. haemocanis (Figure 5, panel B).
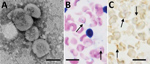
Negative-staining and electron microscopy of Candidatus M. haemohominis particles identified ribosomes, DNA, and soluble RNA in the cytoplasm (Figure 6, panel A). In situ hybridization identified Candidatus M. haemohominis on the surface of erythrocytes and in the cytoplasm of macrophages in bone marrow (Figure 6, panel B).
Monitoring and Treatment for Candidatus M. haemohominis Infection
We determined the level of Candidatus M. haemohominis DNA in serum by using real-time PCR (Figure 7). A high copy number for Candidatus M. haemohominis DNA was detected 18 days after the accidental needlestick injury. The bacterial load in serum decreased below the detection limit (<10 copies/reaction) 14 days after the patient was given levofloxacin, but again increased. Metagenomic analysis also indicated a high detection rate for Mycoplasma spp. reads in serum before and after treatment with levofloxacin. Read-mapping analysis showed that only 2-nt mutations were identified in samples after treatment with levofloxacin. Moreover, 1 of 2 mutations was a nonsynonymous mutation in the quinolone resistance–determining regions (QRDR) of the DNA gyrase subunit A GyrA (i.e., Gly95Cys, corresponding to aa 81 by numbering for Escherichia coli) (Figure 8). The bacterial load promptly decreased after combination therapy with moxifloxacin and minomycin. We also identified Candidatus M. haemohominis by using real-time PCR and in situ hybridization with samples from a liver biopsy of the patient.
Infection with Candidatus M. haemohominis needs to be distinguished from other hemoplasmas in terms of clinical symptoms. As described previously, infection with possible hemoplasmas results in hemolytic anemia and pyrexia (6–11). Our results showed that infection with Candidatus M. haemohominis can cause various life-threatening symptoms in humans, such HPS, liver damage, and bleeding.
We identified Candidatus M. haemohominis in the case-patient and clarified the clinical features of Candidatus M. haemohominis infection as follows. First, Candidatus M. haemohominis infection might cause HPS. Second, this infection might cause reactive plasmacytosis. Third, the increase in plasma cells might induce antibody production and hypergammaglobulinemia. Fourth, infected patients might have coccoid bacterial forms on erythrocyte surfaces and pseudohypoglycemia in vitro. Fifth, this infection might also be accompanied by mild to severe hemorrhagic episodes.
We suggest that our results will help in diagnoses of Candidatus M. haemohominis infections, enabling early therapeutic intervention that might cure patients with these infections. Candidatus M. haemohominis patients are highly susceptible to misdiagnosis with other diseases, such as collagen diseases (e.g., systemic lupus erythematosus) and virus infections (e.g., severe fever with thrombocytopenia syndrome and EBV-associated HPS), because of the wide range of complications.
Generally, antimicrobial drug therapy with fluoroquinolones or tetracyclines is effective against hemoplasmas in animals. Combination therapy (9) or sequential treatment (19) with these drugs can be more effective against hemoplasmas because a single agent is often insufficient for consistent elimination of bacteremia (19). Moreover, detection of mutations in QRDR of gyrase A (Gly95Cys) suggested that Candidatus M. haemohominis is resistant to older fluoroquinolones (e.g., levofloxacin) (20,21). In contrast, newer fluoroquinolones, such as moxifloxacin or sitafloxacin, and tetracyclines, are effective against mycoplasmas harboring this mutation (21).
Laboratory tests for our case-patient showed low blood glucose levels (<5 mg/dL, reference range 61–139 mg/dL) but related no clinical signs. Hemoplasma-associated hypoglycemia in the absence of any associated clinical signs has been described in various animal species (22). This phenomenon is proportional to the severity of bacteremia and depends on the hemoplasma species (22). Because it is possible that glucose would be useful only for carbohydrate metabolism in the hemoplasma species (23), this finding might cause attachment in the epierythrocytic environment, which is rich in glucose, and result in alternative energy source pathways that become redundant.
We observed moderate to severe hemorrhagic episodes for this case-patient. Although the pathogenesis of bleeding in patients infected with Candidatus M. haemohominis is not completely understood, a dramatic decrease in coagulation factor VIII and VWF activities was observed in this case-patient, which is similar to that observed in patients with acquired von Willebrand syndrome (24,25). Given the hypergammaglobulinemia and increased plasma cell levels observed in this case-patient and other patients. the possible mechanisms of infection are adsorption of vWF onto plasma cells or activated platelets apart from the presence of vWF-specific antibodies and inhibitors (24,25).
In conclusion, we identified and characterized Candidatus M. haemohominis infection in a human. Although our study has limitations, our results provide useful knowledge about this infection. Other patients infected with Candidatus M. haemohominis may have died before a confirmed diagnosis was made. Thus, Candidatus M. haemohominis infection might occur more frequently than generally recognized. Further epidemiologic investigations of infection with Candidatus M. haemohominis in humans and of intermediate host(s) for this pathogen would clarify the extent of human infection and possible transmission routes.
Dr Hattori is a research scientist at Showa University School of Medicine, Tokyo, Japan. His primary research interest is hematology.
Acknowledgments
We thank the case-patient for participating in this study, the clinical staff who supported and cared for this patient, and Yuko Sato and Michiyo Kataoka for providing technical assistance with immunohistochemical analysis, in situ hybridization, and electron microscopy.
This study was supported by the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (grants JP19fk0108048, JP19fk0108103, and JP19fk0108104).
N.H., T.T. (Takahiro Takuma), T.I., M.K., T.S., H.K., H.H., T.N., T.W., and Y.N. designed the study and interpreted the data; M.K. performed metagenomic analysis; T.S. performed bioinformatics; H.K. and H.H. performed pathologic analysis; N.A., H.W., N.N., J.E., R.Y., S.I., Y.M., and T.T (Takahiro Tokunaga) provided assistance in the design of the study and characterized patient samples; N.H., M.K., H.K., T.S., and H.H. wrote the manuscript; and T.N., T.W., and Y.N. critically reviewed the manuscript. All authors approved the final version of the manuscript submitted for publication.
References
- Messick JB. Hemotrophic mycoplasmas (hemoplasmas): a review and new insights into pathogenic potential. Vet Clin Pathol. 2004;33:2–13. DOIPubMedGoogle Scholar
- Harasawa R, Fujita H, Kadosaka T, Ando S, Rikihisa Y. Proposal for ‘Candidatus Mycoplasma haemomuris subsp. musculi’ in mice, and ‘Candidatus Mycoplasma haemomuris subsp. ratti’ in rats. Int J Syst Evol Microbiol. 2015;65:734–7. DOIPubMedGoogle Scholar
- Iso T, Suzuki J, Sasaoka F, Sashida H, Watanabe Y, Fujihara M, et al. Hemotropic mycoplasma infection in wild black bears (Ursus thibetanus japonicus). Vet Microbiol. 2013;163:184–9. DOIPubMedGoogle Scholar
- Millán J, López-Roig M, Delicado V, Serra-Cobo J, Esperón F. Widespread infection with hemotropic mycoplasmas in bats in Spain, including a hemoplasma closely related to “Candidatus Mycoplasma hemohominis”. Comp Immunol Microbiol Infect Dis. 2015;39:9–12. DOIPubMedGoogle Scholar
- Sykes JE. Feline hemotropic mycoplasmas. Vet Clin North Am Small Anim Pract. 2010;40:1157–70. DOIPubMedGoogle Scholar
- dos Santos AP, dos Santos RP, Biondo AW, Dora JM, Goldani LZ, de Oliveira ST, et al. Hemoplasma infection in HIV-positive patient, Brazil. Emerg Infect Dis. 2008;14:1922–4. DOIPubMedGoogle Scholar
- Yuan CL, Liang AB, Yao CB, Yang ZB, Zhu JG, Cui L, et al. Prevalence of Mycoplasma suis (Eperythrozoon suis) infection in swine and swine-farm workers in Shanghai, China. Am J Vet Res. 2009;70:890–4. DOIPubMedGoogle Scholar
- Sykes JE, Lindsay LL, Maggi RG, Breitschwerdt EB. Human coinfection with Bartonella henselae and two hemotropic mycoplasma variants resembling Mycoplasma ovis. J Clin Microbiol. 2010;48:3782–5. DOIPubMedGoogle Scholar
- Steer JA, Tasker S, Barker EN, Jensen J, Mitchell J, Stocki T, et al. A novel hemotropic Mycoplasma (hemoplasma) in a patient with hemolytic anemia and pyrexia. Clin Infect Dis. 2011;53:e147–51. DOIPubMedGoogle Scholar
- Maggi RG, Mascarelli PE, Havenga LN, Naidoo V, Breitschwerdt EB. Co-infection with Anaplasma platys, Bartonella henselae and Candidatus Mycoplasma haematoparvum in a veterinarian. Parasit Vectors. 2013;6:103. DOIPubMedGoogle Scholar
- Maggi RG, Compton SM, Trull CL, Mascarelli PE, Mozayeni BR, Breitschwerdt EB. Infection with hemotropic Mycoplasma species in patients with or without extensive arthropod or animal contact. J Clin Microbiol. 2013;51:3237–41. DOIPubMedGoogle Scholar
- Yang D, Tai X, Qiu Y, Yun S. Prevalence of Eperythrozoon spp. infection and congenital eperythrozoonosis in humans in Inner Mongolia, China. Epidemiol Infect. 2000;125:421–6. DOIPubMedGoogle Scholar
- Takeuchi F, Sekizuka T, Yamashita A, Ogasawara Y, Mizuta K, Kuroda M. MePIC, metagenomic pathogen identification for clinical specimens. Jpn J Infect Dis. 2014;67:62–5. DOIPubMedGoogle Scholar
- Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, Mitra S, et al. MEGAN community edition: interactive exploration and analysis of large-scale microbiome sequencing data. PLOS Comput Biol. 2016;12:
e1004957 . DOIPubMedGoogle Scholar - Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. DOIPubMedGoogle Scholar
- Tasker S, Peters IR, Mumford AD, Day MJ, Gruffydd-Jones TJ, Day S, et al. Investigation of human haemotropic Mycoplasma infections using a novel generic haemoplasma qPCR assay on blood samples and blood smears. J Med Microbiol. 2010;59:1285–92. DOIPubMedGoogle Scholar
- Murray GL, Bradshaw CS, Bissessor M, Danielewski J, Garland SM, Jensen JS, et al. Increased macrolide and fluoroquinolone resistance in Mycoplasma genitalium. Emerg Infect Dis. 2017;23:809–12. DOIPubMedGoogle Scholar
- Fricke WF, Wright MS, Lindell AH, Harkins DM, Baker-Austin C, Ravel J, et al. Insights into the environmental resistance gene pool from the genome sequence of the multidrug-resistant environmental isolate Escherichia coli SMS-3-5. J Bacteriol. 2008;190:6779–94. DOIPubMedGoogle Scholar
- Novacco M, Sugiarto S, Willi B, Baumann J, Spiri AM, Oestmann A, et al. Consecutive antibiotic treatment with doxycycline and marbofloxacin clears bacteremia in Mycoplasma haemofelis-infected cats. Vet Microbiol. 2018;217:112–20. DOIPubMedGoogle Scholar
- Hamasuna R, Le PT, Kutsuna S, Furubayashi K, Matsumoto M, Ohmagari N, et al. Mutations in ParC and GyrA of moxifloxacin-resistant and susceptible Mycoplasma genitalium strains. PLoS One. 2018;13:
e0198355 . DOIPubMedGoogle Scholar - Weigel LM, Steward CD, Tenover FC. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob Agents Chemother. 1998;42:2661–7. DOIPubMedGoogle Scholar
- Tasker S, Peters IR, Papasouliotis K, Cue SM, Willi B, Hofmann-Lehmann R, et al. Description of outcomes of experimental infection with feline haemoplasmas: copy numbers, haematology, Coombs’ testing and blood glucose concentrations. Vet Microbiol. 2009;139:323–32. DOIPubMedGoogle Scholar
- Barker EN, Darby AC, Helps CR, Peters IR, Heesom KJ, Arthur CJ, et al. Molecular characterization of the uncultivatable hemotropic bacterium Mycoplasma haemofelis. Vet Res (Faisalabad). 2011;42:83. DOIPubMedGoogle Scholar
- Mohri H. Acquired von Willebrand syndrome: features and management. Am J Hematol. 2006;81:616–23. DOIPubMedGoogle Scholar
- Tiede A, Rand JH, Budde U, Ganser A, Federici AB. How I treat the acquired von Willebrand syndrome. Blood. 2011;117:6777–85. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: December 10, 2019
1These authors contributed equally to this article.
Table of Contents – Volume 26, Number 1—January 2020
| EID Search Options |
|---|
|
|
|
|
|
|




Please use the form below to submit correspondence to the authors or contact them at the following address:
Norimichi Hattori, Division of Hematology, Department of Medicine, Showa University School of Medicine, 1-5-8 Hatanodai, Shinagawa-Ku, Tokyo 142-8666, Japan
Top