Foodborne Origin and Local and Global Spread of Staphylococcus saprophyticus Causing Human Urinary Tract Infections
Opeyemi U. Lawal, Maria J. Fraqueza, Ons Bouchami, Peder Worning, Mette D. Bartels, Maria L. Gonçalves, Paulo Paixão, Elsa Gonçalves, Cristina Toscano, Joanna Empel, Małgorzata Urbaś, M. Angeles Domínguez, Henrik Westh, Hermínia de Lencastre, and Maria Miragaia

Author affiliations: Universidade Nova de Lisboa, Oeiras, Portugal (O.U. Lawal, O. Bouchami, H. de Lencastre, M. Miragaia); Centre for Interdisciplinary Research in Animal Health (CIISA), Universidade de Lisboa, Lisbon, Portugal (M.J., Fraqueza); Hvidovre University Hospital, Hvidovre, Denmark (P. Worning, M.D. Bartels, H. Westh); SAMS Hospital, Lisbon (M.L. Gonçalves); Hospital da Luz, Lisbon (P. Paixão); Hospital Egas Moniz, Lisbon (E. Gonçalves, C. Toscano); Narodowy Instytut Leków, Warsaw, Poland (J. Empel, M. Urbaś); Hospital Universitari de Bellvitge, Barcelona, Spain (M.A. Domínguez); University of Copenhagen, Copenhagen, Denmark (H. Westh); The Rockefeller University, New York, New York, USA (H. de Lencastre)
Main Article
Figure 3
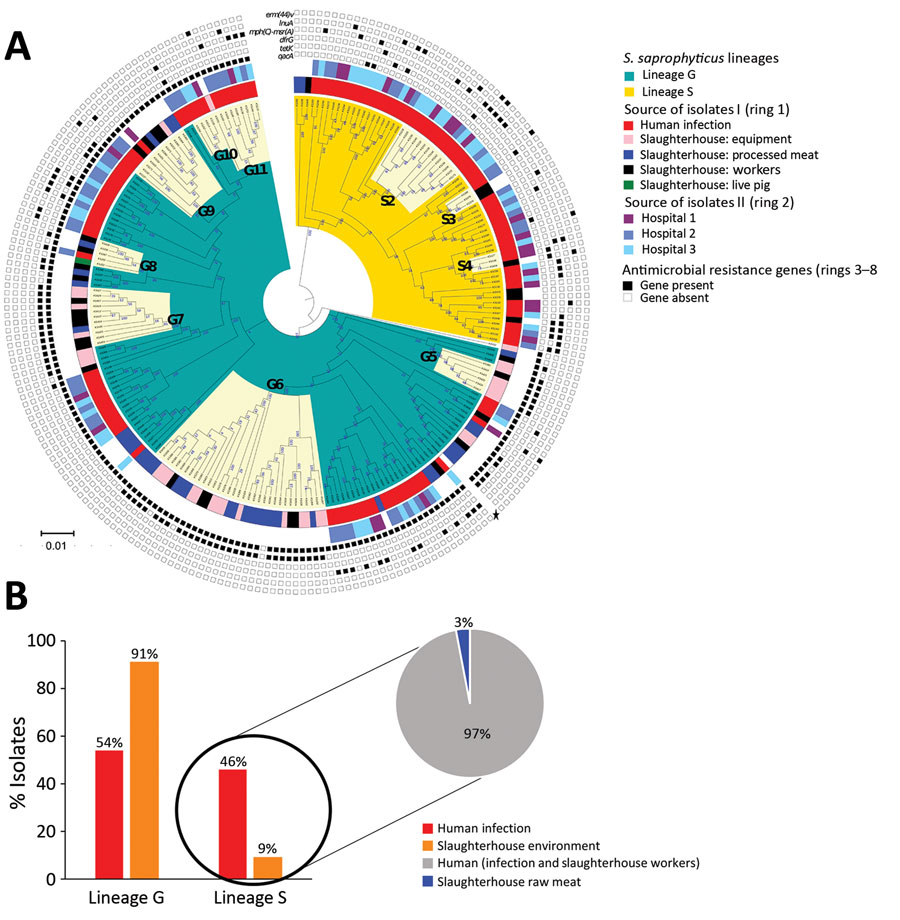
Figure 3. Maximum-likelihood tree depicting the genetic relatedness of Staphylococcus saprophyticus isolates belonging to clonal lineage G recovered from human infections or slaughterhouse contamination, Portugal, 2016–2017. Each node represents a strain. The tree was visualized by using Interactive Tree of Life (iTOL; https://itol.embl.de). Clusters highlighted in cream in the innermost ring represent admixture of isolates in clusters G5–G11, which were recovered from different sources and are closely related by single-nucleotide polymorphism. Ring 1 represents genetic relatedness of isolates recovered from different sites in the slaughterhouses and those recovered from infection in the community. Ring 2 depicts the isolates recovered from different hospitals. Scale bar indicates nucleotide substitutions per site.
Main Article
Page created: December 29, 2020
Page updated: February 21, 2021
Page reviewed: February 21, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
