Volume 28, Number 3—March 2022
Research
Case–Control Study of Clostridium innocuum Infection, Taiwan
Cite This Article
Citation for Media
Abstract
Vancomycin-resistant Clostridium innocuum was recently identified as an etiologic agent for antibiotic-associated diarrhea in humans. We conducted a case–control study involving 152 C. innocuum-infected patients during 2014–2019 in Taiwan, using 304 cases of Clostridioides difficile infection (CDI) matched by diagnosis year, age (+2 years), and sex as controls. The baseline characteristics were similar between the 2 groups. C. innocuum–infected patients experienced more extraintestinal clostridial infection and gastrointestinal tract–related complications than did patients with CDI. The 30-day mortality rate among C. innocuum–infected patients was 14.5%, and the overall rate was 23.0%. Chronic kidney disease, solid tumor, intensive care unit admission, and shock status were 4 independent risk factors for death. C. innocuum identified from clinical specimens should be recognized as a pathogen requiring treatment, and because of its intrinsic vancomycin resistance, precise identification is necessary to guide appropriate and timely antimicrobial therapy.
Clostridium species are obligate anaerobic, endospore-forming bacilli that usually colonize in the gastrointestinal tracts of humans. Of the >200 species of Clostridium, >30 are potential pathogens in humans, such as C. perfringens and Clostridioides difficile. However, C. innocuum has rarely been described as associated with human disease.
C. innocuum was first identified in the 1960s among 8 patients in the United States; the name, innocuum, described its lack of virulence (1,2). It was challenging to distinguish C. innocuum from other Clostridium species (especially C. ramosum and C. clostridioforme, together called the RIC group) because of their similar phenotypes of atypical clostridial colonial morphology, rare spore-forming features, and fatty acid pattern (3–5). Identifying C. innocuum has become faster and more accurate after the introduction of molecular techniques such as 16S RNA sequencing and matrix-associated laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (6).
In 1995, Cutrona et al. reported the first case of endocarditis caused by C. innocuum (7). Although the bacterium was considered less pathogenic and seldom caused infections previously, more and more clinical evidence has emerged since 2000s, suggesting C. innocuum might be a potential cause of antibiotic-associated diarrhea and of extraintestinal clostridial infection (EICI), such as bacteremia, intra-abdominal infection, and endocarditis (8–10). However, we are not aware of a study of C. innocuum infection with a large enough cohort of patients to describe its clinical characteristics.
Precise diagnosis of C. innocuum is necessary because of its unique intrinsic resistance to vancomycin, presumably caused by the presence of 2 chromosomal genes that enable the synthesis of a peptidoglycan precursor terminating in serine with low vancomycin affinity (9,11). Although vancomycin is one of the recommended antimicrobial drugs to treat infections caused by Clostridium species, especially C. difficile, intrinsic resistance to vancomycin in C. innocuum poses the risk for inappropriate treatment for patients who acquire C. innocuum infection (12). C. difficile is one of the most representative clostridial species to cause human disease and has been well investigated. In the United States, ≈500,000 infections were identified annually, and 15,000–30,000 deaths were associated with C. difficile infection (CDI) (12–14)
In previous studies, we demonstrated C. innocuum as a potential invasive pathogen causing severe colitis and EICI in a small case series and proved its cellular toxicity in vitro (8,9). Herein, we conducted a retrospective case–control study to describe and evaluate the clinical characteristics and outcomes of infections caused by C. innocuum. To this end, we selected case-patients with CDI as the control group.
Institute Review Boards in Chang Gung Memorial Hospital (CGMH; Taoyuan, Taiwan) approved the study, allowing review of the medical data of the patients (IRB#201900906B0). A waiver of consent was granted given the retrospective nature of the project and anonymous analysis of the clinical information of patients.
Study Design, Clinical Setting, and Case Enrollment
We conducted a retrospective case–control study at CGMH during 2014–2019. CGMH is a tertiary medical center accommodating 3,700 patient beds. We selected C. difficile as the control to better illustrate the clinical features of C. innocuum infection. The case and control groups were assigned in a 1:2 ratio and matched in the diagnosed year, age +2 years, and sex.
We identified cases with C. innocuum and C. difficile infections using the rapid ID 32A system (bioMérieux, https://www.biomerieux.com) and MALDI-TOF mass spectrometry Biotyper (Bruker Daltonik GmbH, https://www.bruker.com) (15–17). MALDI-TOF mass spectrometry was introduced in 2009 in the clinical microbiology laboratory of CGMH, but C. innocuum was not reported routinely because it was considered a clinically insignificant microorganism. To trace the cases infected with C. innocuum, we reviewed the original reporting database from the MALDI-TOF mass spectrometry system directly and identified the samples reporting C. innocuum. Our definition of a microbiologically confirmed C. innocuum infection was that the original report from the MALDI-TOF mass spectrometry database revealed C. innocuum in the strongest 2 signals and had signal scores >2.00. We defined C. difficile infections by the same rationale.
We reviewed baseline information of each patient and enlisted all patients with C. innocuum infection in the study. We defined C. difficile infection as a positive PCR-based toxin assay with presence of clinical symptoms compatible with the infection, or a positive culture of C. difficile with compatible clinical symptoms (e.g., documented diarrhea or radiologic features of toxic megacolon). We excluded cases with concomitant C. innocuum and C. difficile isolated from the same clinical sample from the study. For the case-control matching, 3 authors (Y.-C. Chen, Y.-C. Kuo, and M.-C. Chen) reviewed baseline information of all cases with C. innocuum and C. difficile infection. We randomly selected 2 controls for each case, matched by diagnostic year, age (+2 years), and sex of the index case. If no controls were eligible from these 3 matching variables, then we dropped the sex criterium, followed by the age criterium if necessary. After the matching process, we further reviewed the clinical information of these patients.
Clinical Data Resources, Variables, and Definition
We collected demographic data, clinical manifestations, laboratory testing results, images, and microbiology reports through an electronic medical record system (EMR). Demographic data were age, sex, race, underlying systemic diseases, and acquisition modality (community vs. hospital). We defined hospital-acquired infection as the symptoms that occurred >48 hours after admission, or <4 weeks after discharge from a healthcare facility; otherwise, it was classified as a community-acquired infection (18). We calculated the Charlson Comorbidity Index score for each patient to represent the baseline physiologic condition affected by underlying disease. The index is composed of 19 underlying conditions in 4 categories. Each category had a weighted score based on the risk for 1- and 10-year mortality rate (19).
We recorded clinical symptoms such as diarrhea, fever, bloody stool, abdominal pain, vomiting, and abdominal distension. We also reviewed disease-related complications, including toxic megacolon, ileus, bowel perforation, and shock. Recurrent infection was defined if the patient had a repeated microbiological culture from the same specimen source within 8 weeks of initial documented symptoms resolution (20,21). Outcome assessment included 30-day, 90-day, and overall deaths after the infection. We reviewed previous antibiotic exposures according to each class: penicillins, cephalosporins, carbapenems, fluoroquinolones, aminoglycosides, macrolides, tetracyclines, glycopeptides, oxazolids, polymyxins, lincosamides, and metronidazole. We defined antibiotic exposure rates as the percentage of patients who received any drugs <30 days before C. innocuum or C. difficile infection and duration of antibiotic exposure as total days of any antimicrobial drug use in a patient <30 days before the event of C. innocuum or C. difficile infection.
Bacterial Isolation and Identification
We performed anaerobic bacterial cultures in the clinical microbiology laboratory, as described previously (9). We streaked all the anaerobic samples onto the selective agar plate, including CDC-ANA-BAP (anaerobic blood agar plate), CDC-ANA-PEA (anaerobic phenylethyl alcohol blood agar plate), and BBE/KVLB (Bacteroides bile esculin and laked kanamycin) bi-plate. We incubated agar plates in anaerobic conditions (90% N2/10% CO2) at 37°C for 5 days. We grossly reviewed the growing colonies on plate and analyzed 1 representative colony for each agar plate by the rapid ID 32A system (bioMérieux) for identification of the microorganisms.
Antimicrobial Susceptibility Testing
We tested antimicrobial susceptibilities to clindamycin, metronidazole, penicillin, piperacillin, and ampicillin/sulbactam by the break-point agar dilution method according to Clinical and Laboratory Standards Institute criteria (document M11-A8) for anaerobic bacteria (22). We used interpretive criteria in document M100S to determine susceptibility (22).
Statistical Analysis
We performed statistical analysis by SPSS Statistics 24.0 (SPSS Inc., https://www.ibm.com/products/spss-statistics). For continuous variables, we determined significance by using the independent t test or Mann-Whitney U test as appropriate. If the continuous variable had outliers and did not fit the normal distribution, variables were shown as median (interquartile range, range). We analyzed the categorical variables by χ2 test and considered p<0.05 statistically significant. We obtained odds ratios (ORs) from cross-tabulation and analyzed the p value of ORs by univariate logistic regression. We estimated mortality rate at 30 days and 90 days after the positive culture and analyzed by Kaplan-Meier survival analysis using methods described previously (23). In addition, we examined risk factors associated with 30- and 90-day mortality in both groups by logistic regression.
Participants and Demographic Information
By the MALDI-TOF mass spectrometry system, 180 samples yielded the growth of C. innocuum. We excluded 22 of those from further analysis because of lack of access to clinical information and 6 because of concomitant isolation of C. innocuum and C. difficile from the same sample (CI group). We matched the control group with C. innocuum samples in accordance with the study criteria. From 1,134 C. difficile cases during the study period, we enrolled 304 cases as controls (CD group). All control cases were matched precisely on diagnostic year and age (+2 years); 25 controls were not matched on sex. The mean patient age for the 456 cases was 66.7 years, and 58.3% of patients were male (Table 1). Both groups were similar regarding age, sex, and Charlson Comorbidity Index score (5.7 + 3.2 for CI and 6.2 + 3.3 for CD). Subgroup analysis of each age group (<50, 50–60, 60–70, 70–80, and >80 years) also revealed no statistical difference. Overall, 8 pediatric patients were recruited, 3 in the CI group and 5 in the CD group. Regarding underlying systemic diseases, the CD group showed more patients with chronic kidney disease (18.4% vs. 30.9%; p = 0.005) (Table 1). Of note, more patients acquired the infection in the community in the CI group (33.6% vs. 16.8%; odds ratio [OR] 2.5, 95% CI 1.6–3.9; p<0.001) (Table 1).
Disease Characteristics and Severity
We observed notable differences in disease characteristics between the 2 groups. Those in the CI group had a 6.5 times higher risk of developing EICI, including bacteremia, intra-abdominal infection, biliary tract infection, skin and soft tissue infection, pyospermia, and bacterial vaginitis (36.8% for CI vs. 8.2% for CD; OR 6.5, 95% CI 3.9 –11.0; p<0.001) (Table 1). On the contrary, most disease manifestation in the CD group was confined to the intestine and colon, mainly C. difficile–associated diarrhea. Most patient had antibiotic exposure 30 days before the CI or CD infection event. CD group showed higher 30-day antibiotic exposure rate (95.1%) than CI group (79.6%; p<0.001) (Table 2) and longer duration (mean 15.6 days, SD 8.3) than CI group (mean 13.7 days, SD 8.6; p<0.001). Patients in CD group received more penicillins, cephalosporins, carbapenems, and fluroquinolones (Table 2).
Regarding disease severity, most of the patients in both groups required hospitalization (93.4% in the CI group and 97.7% in the CD group; p = 0.03) (Table 1). Although most patients in CD group had intestinal infections, gastrointestinal tract–related complications of ileus, bowel perforation, clinical sepsis, and shock occurred more frequently in the CI group (26.3%) than CD group (11.2%; OR 2.8, 95% CI 1.7–4.7; p<0.001). CI group also showed a higher rate of intensive care unit (ICU) admission (23.6% vs. 9.5%; OR 2.9, 95% CI 1.7–5.0; p<0.001) (Table 1). All the data indicated that the disease severity at the acute stage was more severe and invasive in the C. innocuum–infected patients. Furthermore, we saw no recurrence of infection in CI group but recurrence of infection in 4.9% of CD group (p = 0.005).
We observed no statistically significant differences in clinical presentations, but patients with C. innocuum infection had fewer diarrheal symptoms and less fever. In the laboratory testing results, patients experienced anemia more commonly in the CD group than CI group; hemoglobin counts were 9.8 (2.0) g/dL in CD and 10.7 (2.4) g/dL in CI (p<0.001) (Table 3). We observed no difference in other systemic inflammatory markers. A limited number of patients received colonoscopy examination, and we found no pseudomembranous colitis in the CI group.
Outcome and Risk Factor for Mortality Rate
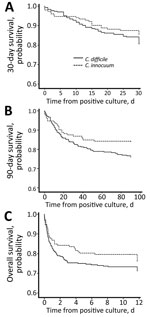
The 30-day mortality rate in the CI group was 14.5%; the 90-day rate, 15.8%, and the overall rate, 23.0%. Although the 90-day mortality rate was slightly higher in the CD group with a significant difference (p value of log rank test = 0.05) in Kaplan-Meier survival analysis, the overall mortality rate did not show a statistically significant difference between the 2 groups (Figure). Using logistic regression, we identified chronic kidney disease (OR 8.6, 95% CI 2.6–28.4; p<0.001), solid tumor (OR 3.5, 95% CI 1.0–12.0; p = 0.051), ICU admission (OR 7.3, 95% CI 2.4–21.9; p<0.001), and shock status (odds ratio 8.0, 95% CI 2.4–27.2; p<0.001) as 4 independent risk factors for both 30-day and overall mortality rates in the patients with C. innocuum infection. We identified 7 bacteremias caused by C. innocuum in this study. Two of those patients experienced septic shock, and 1 needed ICU hospitalization. The 30-day mortality rate for the 7 patients was 42.9% (3/7) and 90-day was 57.1% (4/7).
Microbiologic Result and Antimicrobial Susceptibility
Among the 152 C. innocuum isolates, we recovered 96 (63.2%) isolates from stool specimens; the rest were from the blood (7), ascites (13), pus/abscess (16), wound/deep tissue (16), bile juice (2), endocervix (1), and semen (1). We detected 18 polymicrobial infections in the CI group, most of which were from ascites and pus/abscess samples. More C. innocuum isolates (36.8%) than C. difficile isolates (8.2%) were from extraintestinal specimens (p<0.001) (Table 3), which is compatible with our clinical observation. We performed antimicrobial susceptibility testing on limited isolates. In the C. innocuum isolates, we observed the highest susceptibility rate for metronidazole (20/20, 100%) and ampicillin/sulbactam (21/21, 100%), followed by penicillin (35/44, 79.5%) and clindamycin 30/44 (68.2%).
Genus Clostridium is large and heterogeneous; it includes <200 species. Accurate species identification has been difficult. In recent years, several new species have been recognized and others reclassified using newer molecular diagnostic methods, such as 16S rRNA gene sequencing (24). Among the medically important Clostridium spp., C. perfringens is the predominant species isolated from cases of bacteremia. The severity of EICI varies; for bacteremia, the mortality rate was found to be 48%–52% by different studies (25–27). The risk factors for disease acquisition and death were related to an underlying immunocompromised condition such as hemodialysis, malignancy, immunosuppressant use, and Crohn’s disease (25). The main portal of entry is the hepatobiliary and gastrointestinal tract. We believe this is also the case in C. innocuum because stool was a common source for the C. innocuum isolates and gastrointestinal tract–related complications were not uncommon in C. innocuum–infected patients. A recent study by Ha et al. (28) also found that C. innocuum is one of the most common bacteria that could translocate from intestine to mesenteric tissue in patients with Crohn’s disease and further induce adipogenesis and local fibrosis, known together as creeping fat.
We found that among anaerobic clostridial species, C. innocuum has long been overlooked as a human pathogen. Our study is to date the most comprehensive observational study to depict the clinical manifestations and outcome of C. innocuum infection; not only it is more invasive than most Clostridium species, but it can cause more gastrointestinal tract complications following intestinal infection. Case reports of EICI related to C. innocuum infection have been published from the United States, Spain, Japan, and Taiwan (10,29–32) (Table 4). Bacteremia and intra-abdominal infection were the most common manifestations, which is compatible with our observations. All the infections occurred in patients with underlying conditions; prolonged antimicrobial therapy was required to treat these patients, whose mortality rate (20%) was similar to that observed in our study (23%). Compared to C. difficile, which is known to be a nosocomial pathogen, nearly one third of the C. innocuum infections occurred in the community. This observation indicates that C. innocuum could be more virulent and competitive than C. difficile.
Among the EICI, bacteremia is the most severe form of infection. In a recent study by Morel et al. (33), non–C. difficile Clostridium bacteremia requiring ICU hospitalization showed an aggressive clinical course and was usually life-threatening. The 28-day mortality rate was 55% and the 90-day mortality rate was 71% (33). This report is compatible with our findings of 30-day (42.9%) and 90-day (57.1%) mortality rates in the CI bacteremic patients.
Identifying C. innocuum infection is important because the microorganism expresses intrinsic resistance to vancomycin, because of the synthesis of peptidoglycan precursors with low affinity for vancomycin (MIC 4–16 mg/L) (8,26). Moreover, highly vancomycin-resistant strains (MIC >16 mg/L) could develop if the bacteria were previously exposed to vancomycin (34). Because oral vancomycin has been recommended as the first-line therapy for C. difficile infection, distinguishing C. innocuum from other clostridial species becomes essential to avoid treatment failure caused by inappropriate antimicrobial use. Metronidazole and clindamycin appear to be appropriate choices for treating C. innocuum infection, according to our antimicrobial susceptibility testing results.
The main limitation of our study is the retrospective study design and the inevitable missing data. The lack of standardized medical record format prevented us from precisely defining every case-patient’s diagnosis, especially antibiotic-associated diarrhea and acute colitis, which have similar clinical descriptions in the medical records. Some objective data were not available, which may potentially compromise the accuracy of the estimated rates of presentations and diagnoses among the patients. However, the proportion of missing data appeared small and should not significantly affect the results of the study. Second, not all the C. innocuum isolates from the enrolled patients were tested for antimicrobial susceptibility, and that testing did not include vancomycin. Third, the study does not advance our understanding on virulence mechanism of C. innocuum. It is possible that C. innocuum possesses a unique virulence mechanism to cause gastrointestinal as well as extraintestinal infections, such as the lipopolysaccharide-like structure we described in our previous study (9). C. difficile also contains surface lipocarbohydrate, which has a similar biologic activity to the lipopolysaccharide in gram-negative bacteria (35); this hypothesis needs further experimental verification.
In conclusion, C. innocuum should be considered an important Clostridium species causing EICI and gastrointestinal infection that has a risk for severe complications and a high mortality rate in immunocompromised patients; physicians should recognize it as a pathogen to treat clinically. More studies are needed to understand the virulence mechanism of C. innocuum. Precise identification of C. innocuum will guide appropriate and timely antimicrobial therapy for patients because of its intrinsic vancomycin resistance.
Dr. Chen is an infectious disease specialist at Chang Gung Memorial Hospital, Chang Gung University, Taoyuan, Taiwan. Her research interests are epidemiology, pathogenesis, and resistance mechanisms of enteric bacterial infection.
Acknowledgment
The study was financially supported by grants (CIRPG3H0031-2 and CIRPG3H0041-2) from Chang Gung Memorial Hospital, Taiwan, and the Maintenance Project of the Center for Big Data Analytics and Statistics at Chang Gung Memorial Hospital (grant CLRPG3D0048) for statistical consultation and data analysis.
References
- Smith LD, King E. Clostridium innocuum, sp. n., a sporeforming anaerobe isolated from human infections. J Bacteriol. 1962;83:938–9. DOIPubMedGoogle Scholar
- Alexander CJ, Citron DM, Brazier JS, Goldstein EJ. Identification and antimicrobial resistance patterns of clinical isolates of Clostridium clostridioforme, Clostridium innocuum, and Clostridium ramosum compared with those of clinical isolates of Clostridium perfringens. J Clin Microbiol. 1995;33:3209–15. DOIPubMedGoogle Scholar
- Stokes NA, Hylemon PB. Characterization of delta 4-3-ketosteroid-5 beta-reductase and 3 beta-hydroxysteroid dehydrogenase in cell extracts of Clostridium innocuum. Biochim Biophys Acta. 1985;836:255–61. DOIPubMedGoogle Scholar
- Carlier JP, Sellier N. Identification by gas chromatography-mass spectrometry of short-chain hydroxy acids produced by Fusobacterium species and Clostridium innocuum. J Chromatogr A. 1987;420:121–8. DOIPubMedGoogle Scholar
- Johnston NC, Goldfine H, Fischer W. Novel polar lipid composition of Clostridium innocuum as the basis for an assessment of its taxonomic status. Microbiology (Reading). 1994;140:105–11. DOIPubMedGoogle Scholar
- Li Y, Shan M, Zhu Z, Mao X, Yan M, Chen Y, et al. Application of MALDI-TOF MS to rapid identification of anaerobic bacteria. BMC Infect Dis. 2019;19:941. DOIPubMedGoogle Scholar
- Cutrona AF, Watanakunakorn C, Schaub CR, Jagetia A. Clostridium innocuum endocarditis. Clin Infect Dis. 1995;21:1306–7. DOIPubMedGoogle Scholar
- Chia JH, Wu TS, Wu TL, Chen CL, Chuang CH, Su LH, et al. Clostridium innocuum is a vancomycin-resistant pathogen that may cause antibiotic-associated diarrhoea. Clin Microbiol Infect. 2018;24:1195–9. DOIPubMedGoogle Scholar
- Chia JH, Feng Y, Su LH, Wu TL, Chen CL, Liang YH, et al. Clostridium innocuum is a significant vancomycin-resistant pathogen for extraintestinal clostridial infection. Clin Microbiol Infect. 2017;23:560–6. DOIPubMedGoogle Scholar
- Castiglioni B, Gautam A, Citron DM, Pasculle W, Goldstein EJC, Strollo D, et al. Clostridium innocuum bacteremia secondary to infected hematoma with gas formation in a kidney transplant recipient. Transpl Infect Dis. 2003;5:199–202. DOIPubMedGoogle Scholar
- David V, Bozdogan B, Mainardi JL, Legrand R, Gutmann L, Leclercq R. Mechanism of intrinsic resistance to vancomycin in Clostridium innocuum NCIB 10674. J Bacteriol. 2004;186:3415–22. DOIPubMedGoogle Scholar
- Peng Z, Ling L, Stratton CW, Li C, Polage CR, Wu B, et al. Advances in the diagnosis and treatment of Clostridium difficile infections. Emerg Microbes Infect. 2018;7:15. DOIPubMedGoogle Scholar
- Jewkes J, Larson HE, Price AB, Sanderson PJ, Davies HA. Aetiology of acute diarrhoea in adults. Gut. 1981;22:388–92. DOIPubMedGoogle Scholar
- McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis. 2018;66:987–94. DOIPubMedGoogle Scholar
- Shannon S, Kronemann D, Patel R, Schuetz AN. Routine use of MALDI-TOF MS for anaerobic bacterial identification in clinical microbiology. Anaerobe. 2018;54:191–6. DOIPubMedGoogle Scholar
- Veloo AC, de Vries ED, Jean-Pierre H, Justesen US, Morris T, Urban E, et al.; ENRIA workgroup. The optimization and validation of the Biotyper MALDI-TOF MS database for the identification of Gram-positive anaerobic cocci. Clin Microbiol Infect. 2016;22:793–8. DOIPubMedGoogle Scholar
- Li Y, Shan M, Zhu Z, Mao X, Yan M, Chen Y, et al. Application of MALDI-TOF MS to rapid identification of anaerobic bacteria. BMC Infect Dis. 2019;19:941. DOIPubMedGoogle Scholar
- Khanna S, Pardi DS, Aronson SL, Kammer PP, Orenstein R, St Sauver JL, et al. The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am J Gastroenterol. 2012;107:89–95. DOIPubMedGoogle Scholar
- Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–83. DOIPubMedGoogle Scholar
- Pepin J, Alary ME, Valiquette L, Raiche E, Ruel J, Fulop K, et al. Increasing risk of relapse after treatment of Clostridium difficile colitis in Quebec, Canada. Clin Infect Dis. 2005;40:1591–7. DOIPubMedGoogle Scholar
- Lee HY, Hsiao HL, Chia CY, Cheng CW, Tsai TC, Deng ST, et al. Risk factors and outcomes of Clostridium difficile infection in hospitalized patients. Biomed J. 2019;42:99–106. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing: 27th edition (M100–S27). Wayne (PA): The Institute; 2017.
- Van Daele E, Van de Putte D, Ceelen W, Van Nieuwenhove Y, Pattyn P. Risk factors and consequences of anastomotic leakage after Ivor Lewis oesophagectomy. Interact Cardiovasc Thorac Surg. 2016;22:32–7. DOIPubMedGoogle Scholar
- Finegold SM, Song Y, Liu C, Hecht DW, Summanen P, Könönen E, et al. Clostridium clostridioforme: a mixture of three clinically important species. Eur J Clin Microbiol Infect Dis. 2005;24:319–24. DOIPubMedGoogle Scholar
- Leal J, Gregson DB, Ross T, Church DL, Laupland KB. Epidemiology of Clostridium species bacteremia in Calgary, Canada, 2000-2006. J Infect. 2008;57:198–203. DOIPubMedGoogle Scholar
- Bodey GP, Rodriguez S, Fainstein V, Elting LS. Clostridial bacteremia in cancer patients. A 12-year experience. Cancer. 1991;67:1928–42. DOIPubMedGoogle Scholar
- Shah M, Bishburg E, Baran DA, Chan T. Epidemiology and outcomes of clostridial bacteremia at a tertiary-care institution. ScientificWorldJournal. 2009;9:144–8. DOIPubMedGoogle Scholar
- Ha CWY, Martin A, Sepich-Poore GD, Shi B, Wang Y, Gouin K, et al. Translocation of viable gut microbiota to mesenteric adipose drives formation of creeping fat in humans. Cell. 2020;183:666–683.e17. DOIPubMedGoogle Scholar
- Crum-Cianflone N. Clostridium innocuum Bacteremia in a patient with acquired immunodeficiency syndrome. Am J Med Sci. 2009;337:480–2. DOIPubMedGoogle Scholar
- Hung YP, Lin HJ, Wu CJ, Chen PL, Lee JC, Liu HC, et al. Vancomycin-resistant Clostridium innocuum bacteremia following oral vancomycin for Clostridium difficile infection. Anaerobe. 2014;30:24–6. DOIPubMedGoogle Scholar
- Mutoh Y, Hirai R, Tanimura A, Matono T, Morino E, Kutsuna S, et al. Osteomyelitis due to Clostridium innocuum in a patient with acute lymphoblastic leukemia: case report and literature review. Springerplus. 2015;4:385. DOIPubMedGoogle Scholar
- Aroca-Ferri M, Suárez-Hormiga L, Bosch-Benitez-Parodi E, Bolaños-Rivero M. Peritonitis by Clostridium innocuum associated to peritoneal dialysis [in Spanish]. Rev Esp Quimioter. 2019;32:192–3.
- Morel G, Mulier G, Ghrenassia E, Abdel Nabey M, Tandjaoui Y, Kouatchet A, et al. Non–C. difficile Clostridioides bacteremia in intensive care patients, France. Emerg Infect Dis. 2021;27:1840–9. DOIPubMedGoogle Scholar
- Cherny KE, Ozer EA, Kochan TJ, Johnson S, Kociolek LK. Complete genome sequence of Clostridium innocuum strain LC–LUMC-CI-001, isolated from a patient with recurrent antibiotic-associated diarrhea. Microbiol Resour Announc. 2020;9:e00365–20. DOIPubMedGoogle Scholar
- Sánchez-Hurtado K, Poxton IR. Enhancement of the cytotoxic activity of Clostridium difficile toxin A by surface-associated antigens. J Med Microbiol. 2008;57:739–44. DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleOriginal Publication Date: February 04, 2022
1These authors contributed equally to this article.
Table of Contents – Volume 28, Number 3—March 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Cheng-Hsun Chiu, Chang Gung Children’s Hospital—Department of Pediatrics, 5 Fu-Hsin St, Kweishan Taoyuan County 333, Taiwan
Top