Volume 29, Number 8—August 2023
Research
Prospecting for Zoonotic Pathogens by Using Targeted DNA Enrichment
Figure 1
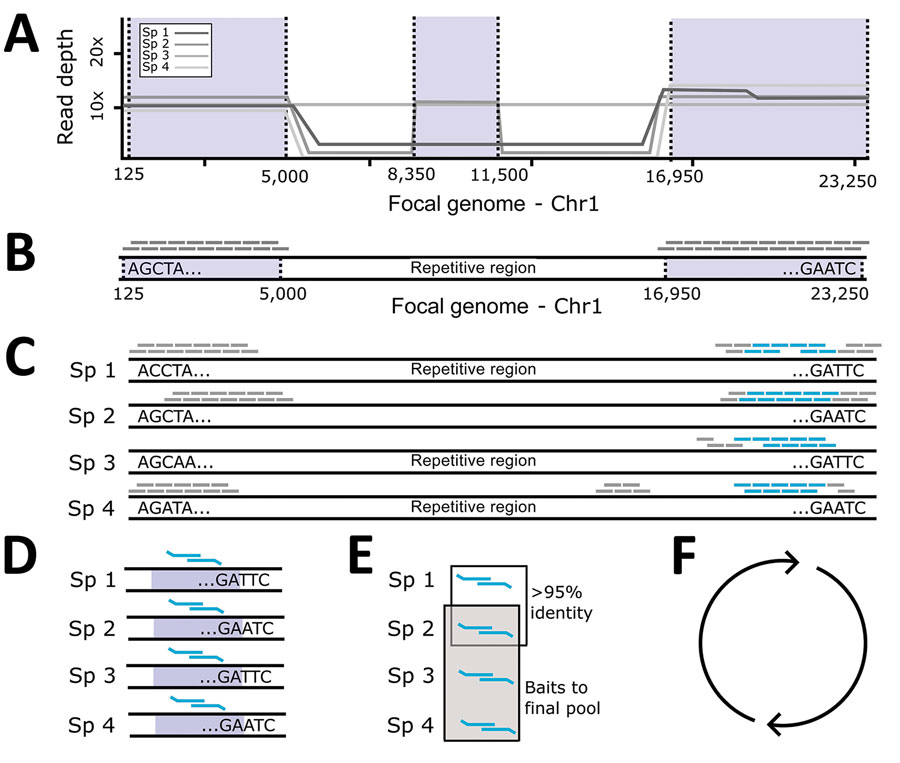
Figure 1. Probe panel design for study of prospecting for zoonotic pathogens by using targeted DNA enrichment. A) Simulated reads from each pathogen within a group were mapped back to a single focal genome. B) We identified regions with consistent coverage from each member of the pathogen group to identify putative, orthologous loci and generated a set of in silico probes from the focal genome. C) Those in silico probes were then mapped back to the genomes of each member in the pathogen group to find single copy, orthologous regions, present in most members. D, E) We designed 2 overlapping 80-bp baits to target the loci in each member of the pathogen group (D) and compared them with each another to remove highly similar probes (E). One probe was retained from each group of probes with high sequence similarity (>95%). F) We identified the probes necessary to capture 49 loci in that pathogen group. This process was repeated for the next pathogen group. Finally, all probes were combined together into a single panel. Chr, chromosome; Sp, specimen.