Volume 31, Number 1—January 2025
Research
Trichuriasis in Human Patients from Côte d’Ivoire Caused by Novel Species Trichuris incognita with Low Sensitivity to Albendazole/Ivermectin Combination Treatment
Figure 2
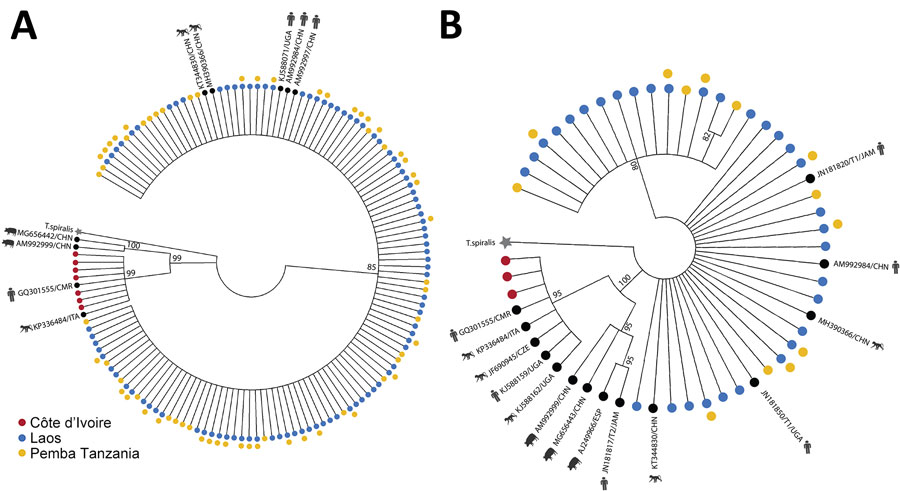
Figure 2. Phylogenetic analysis of ITS1 and ITS2 rRNA ASVs in study of novel Trichuris incognita identified in patient fecal samples from Côte d’Ivoire. Maximum-likelihood method was used to construct trees for Trichuris ITS1 (A) and ITS2 (B) ASVs from Côte d’Ivoire, Laos, and Pemba Island, Tanzania, as well as additional Trichuris reference sequences from pigs, humans, and nonhuman primates in GenBank. Trees were generated by amplicon sequencing of fecal sample DNA from patients in Côte d’Ivoire, Laos, and Pemba Island, Tanzania. After generating the clusters, trees were transformed into cladograms for visualization. Trichinella spiralis (GenBank accession no. KC006432) was used as the outgroup. Each tip of the tree is an ASV or a sequence from GenBank. Colors indicate the 3 regions. Black circles indicate GenBank sequences. Kimura 80 model for ITS-1 and general time reversible model for ITS-1 were chosen as the best nucleotide substitution models. Models were chosen by using jModeltest version 2.1.10 (https://github.com/ddarriba/jmodeltest2). Trees were constructed by using PhyML v3.3 (https://github.com/stephaneguindon/phyml) with 100 bootstrap replicates. Trees were condensed by using MEGA version 11 (https://www.megasoftware.net) to only display branches with consensus support >80%. Trees not to scale. ASV, amplicon sequencing variant; ITS, internal transcribed spacer.
1Current affiliation: Kwantlen Polytechnic University, Surrey, British Columbia, Canada.