Trichuriasis in Human Patients from Côte d’Ivoire Caused by Novel Species Trichuris incognita with Low Sensitivity to Albendazole/Ivermectin Combination Treatment
Abhinaya Venkatesan
1, Rebecca Chen, Max Bär, Pierre H.H. Schneeberger, Brenna Reimer, Eveline Hürlimann, Jean T. Coulibaly, Said M. Ali, Somphou Sayasone, John Soghigian, Jennifer Keiser, and John Stuart Gilleard

Author affiliation: University of Calgary, Calgary, Alberta, Canada (A. Venkatesan, R. Chen, B. Reimer, J. Soghigian, J.S. Gilleard); Swiss Tropical and Public Health Institute, Allschwil, Switzerland (M. Bär, P.H.H. Schneeberger, E. Hürlimann, J. Keiser); University of Basel, Basel, Switzerland (M. Bär, P.H.H. Schneeberger, E. Hürlimann, J. Keiser); Université Félix Hophouët-Boigny, Abidjan, Côte d’Ivoire (J.T. Coulibaly); Public Health Laboratory Ivo de Carneri, Chake Chake, Pemba Island, Tanzania (S.M. Ali); Lao Tropical and Public Health Institute, Vientiane, Laos (S. Sayasone)
Main Article
Figure 3
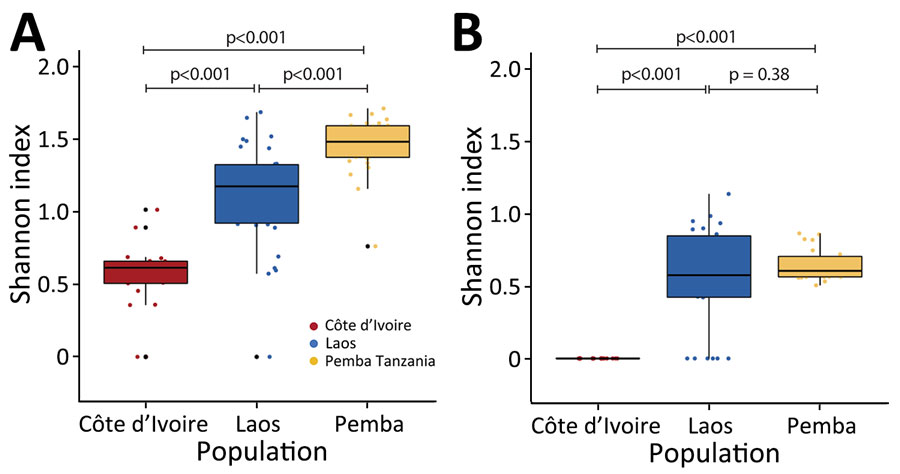
Figure 3. Alpha diversity of ITS1 and ITS2 rRNA amplicon sequence variants in study of novel Trichuris incognita identified in patient fecal samples from Côte d’Ivoire. Shannon-Wiener Index values, indicating α diversity, were determined for Trichuris spp. ITS1 (A) and ITS2 (B) amplicon sequence variants generated by sequencing fecal sample DNA from patients in Cote d’Ivoire, Laos, and Pemba Island, Tanzania. Horizontal lines within boxes indicate medians; box tops and bottoms indicate upper (third) and lower (first) quartiles; error bars (whiskers) indicate minimum and maximum values. Pairwise t-tests were used to determine p values. ITS, internal transcribed spacer.
Main Article
Page created: November 14, 2024
Page updated: December 22, 2024
Page reviewed: December 22, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
