Volume 31, Number 1—January 2025
Research
Trichuriasis in Human Patients from Côte d’Ivoire Caused by Novel Species Trichuris incognita with Low Sensitivity to Albendazole/Ivermectin Combination Treatment
Figure 4
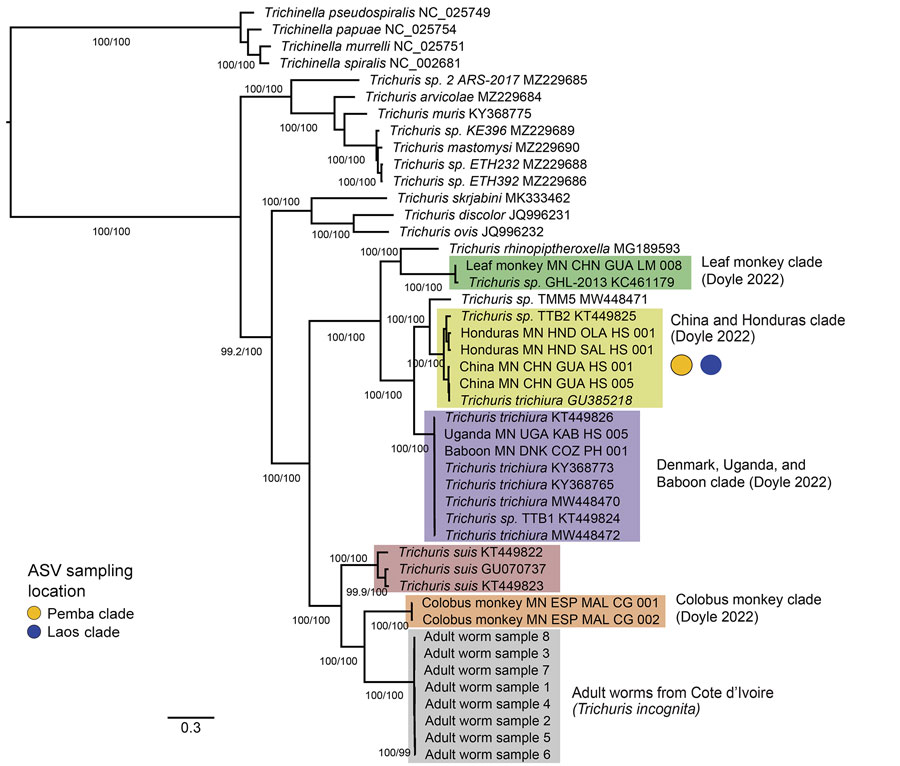
Figure 4. Phylogenetic analysis of Trichuris spp. from complete mitochondrial genome sequences in study of novel Trichuris incognita identified in patient fecal samples from Côte d’Ivoire. Tree was reconstructed by using the maximum-likelihood method for 12 mitochondrial protein coding genes from Trichuris spp. compared with sequences from GenBank. Tree was constructed by using IQ-TREE; alignments were made for the 12 protein coding genes from 45 mitochondrial genomes, including 8 T. incognita sequences obtained from an expulsion study of patients in Côte d’Ivoire (M.A. Bär et al., unpub. data, https://doi.org/10.1101/2024.06.11.598441). Ultrafast bootstrap/Shimodaira-Hasegawa–like branch support values >95/95 are indicated on branches. Color-shaded boxes indicate clades previously identified in the literature. Yellow (Pemba Island) and blue (Laos) colored circles indicate where the clades mapped according to cox-1, nad-1, and nad-4 mitochondrial ASVs. Scale bar indicates nucleotide substitutions per site. ASV, amplicon sequencing variant.
1Current affiliation: Kwantlen Polytechnic University, Surrey, British Columbia, Canada.