Volume 31, Number 4—April 2025
Research
Case–Control Study of Factors Associated with Hemolytic Uremic Syndrome among Shiga Toxin–Producing Escherichia coli Patients, Ireland, 2017–2020
Figure
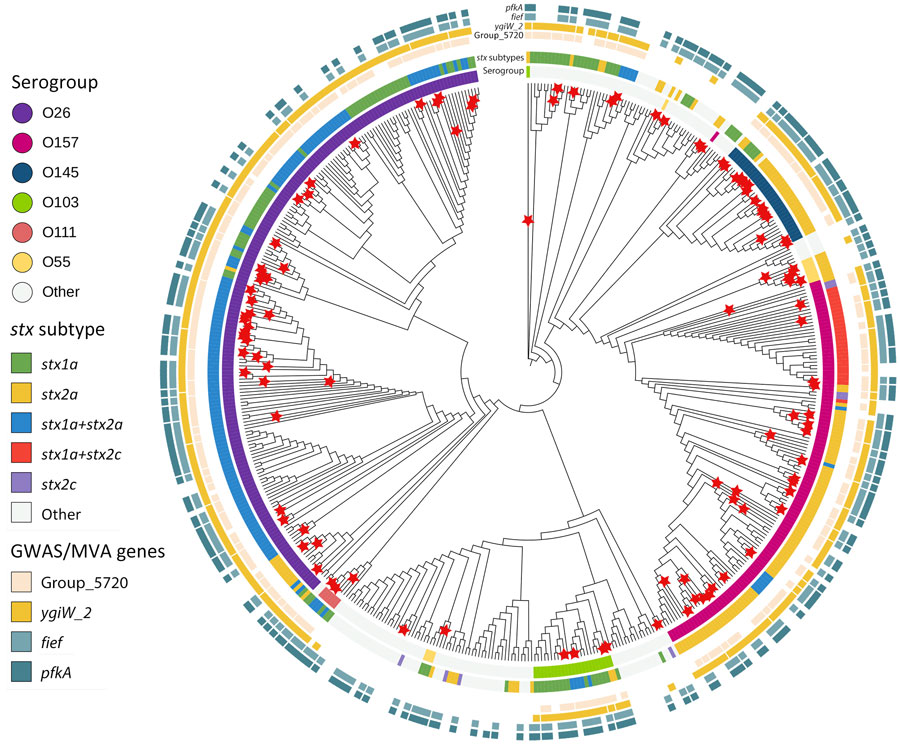
Figure. Maximum-likelihood phylogenetic tree of HUS and non-HUS STEC isolates from study of HUS among patients with STEC, Ireland, 2017–2020. Tree was generated by using RaxML (23) on the basis of a multi-FASTA alignment of the core genes of the 531 STEC isolates. We annotated and visualized the final tree by using iTOL version 6.8.1 (https://itol.embl.de) (24). HUS cases (indicated by red stars) were distributed across several serogroups: O26 (36%), O157 (26%), O145 (14%), O103 (4.6%), O111 (2.8%), and O55 (5.6%). GWAS, genomewide association study; HUS, hemolytic uremic syndrome; MVA, multivariable logistic regression analysis; STEC, Shiga toxin–producing Escherichia coli.
References
- Health Service Executive Health Protection Surveillance Centre. VTEC infection in Ireland, 2017. 2019 [cited 2023 Feb 7]. https://www.hpsc.ie/a-z/gastroenteric/vtec/epidemiologicaldata/annualreportsonepidemiologyofverotoxigenicecoli/VTEC%20infection%20in%20Ireland%202017.pdf
- Health Service Executive Health Protection Surveillance Centre. Infectious intestinal disease: public health & clinical guidance, version 1.1. 2012 [cited 2023 Feb 7]. https://www.hpsc.ie/a-z/gastroenteric/gastroenteritisoriid/guidance/File,13492,en.pdf
- Brzuszkiewicz E, Thürmer A, Schuldes J, Leimbach A, Liesegang H, Meyer FD, et al. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC). Arch Microbiol. 2011;193:883–91. DOIPubMedGoogle Scholar
- Hamilton D, Cullinan J. A practical composite risk score for the development of Haemolytic Uraemic Syndrome from Shiga toxin-producing Escherichia coli. Eur J Public Health. 2019;29:861–8. DOIPubMedGoogle Scholar
- Boerlin P, McEwen SA, Boerlin-Petzold F, Wilson JB, Johnson RP, Gyles CL. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol. 1999;37:497–503. DOIPubMedGoogle Scholar
- Wong CS, Mooney JC, Brandt JR, Staples AO, Jelacic S, Boster DR, et al. Risk factors for the hemolytic uremic syndrome in children infected with Escherichia coli O157:H7: a multivariable analysis. Clin Infect Dis. 2012;55:33–41. DOIPubMedGoogle Scholar
- European Centre for Disease Prevention and Control. Shiga toxin/verocytotoxin–producing Escherichia coli (STEC/VTEC) infection annual epidemiological report for 2018. Stockholm; The Centre; 2020.
- Rice T, Quinn N, Sleator RD, Lucey B. Changing diagnostic methods and increased detection of verotoxigenic Escherichia coli, Ireland. Emerg Infect Dis. 2016;22:1656–7. DOIPubMedGoogle Scholar
- HSE Health Protection Surveillance Centre. Case definitions 2019 [cited 2024 May 7]. https://www.hpsc.ie/a-z/gastroenteric/vtec/casedefinitions
- European Union. Commission implementing decision (EU) 2018/945 of 22 June 2018 on the communicable diseases and related special health issues to be covered by epidemiological surveillance as well as relevant case definitions. 2018 [cited 2024 Aug 6]. https://eur-lex.europa.eu/eli/dec_impl/2018/945/oj
- Ethelberg S, Olsen KEP, Scheutz F, Jensen C, Schiellerup P, Enberg J, et al. Virulence factors for hemolytic uremic syndrome, Denmark. Emerg Infect Dis. 2004;10:842–7. DOIPubMedGoogle Scholar
- HSE Health Protection Surveillance Centre. VTEC enhanced surveillance form CIDR EVENT ID HSE ID 2018 [cited 2024 May 7]. https://www.hpsc.ie/a-z/gastroenteric/vtec/surveillanceinvestigativeforms/VTEC_EnhancedQs2018_v2.0 FINAL_20180219.pdf.
- Department of Health and Children. Health statistics 2002 LENUS (Irish Health Repository). Dublin; 2003 [cited 2023 Jan 18]. https://www.lenus.ie/handle/10147/241861
- Yang X, Sun H, Fan R, Fu S, Zhang J, Matussek A, et al. Genetic diversity of the intimin gene (eae) in non-O157 Shiga toxin-producing Escherichia coli strains in China. Sci Rep. 2020;10:3275. DOIPubMedGoogle Scholar
- Zhang WL, Köhler B, Oswald E, Beutin L, Karch H, Morabito S, et al. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J Clin Microbiol. 2002;40:4486–92. DOIPubMedGoogle Scholar
- Ooka T, Seto K, Kawano K, Kobayashi H, Etoh Y, Ichihara S, et al. Clinical significance of Escherichia albertii. Emerg Infect Dis. 2012;18:488–92. DOIPubMedGoogle Scholar
- Afgan E, Nekrutenko A, Grüning BA, Blankenberg D, Goecks J, Schatz MC, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022;50(W1):
W345–51 . - Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. DOIPubMedGoogle Scholar
- Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016;17:1–9. DOIGoogle Scholar
- Bateman A, Martin MJ, Orchard S, Magrane M, Ahmad S, Alpi E, et al.; UniProt Consortium. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023;51(D1):D523–31. DOIPubMedGoogle Scholar
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–12. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. DOIPubMedGoogle Scholar
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–6. DOIPubMedGoogle Scholar
- Fox J, Weisberg S. An R companion to applied regression. 3rd ed. Thousand Oaks (CA): Sage Publications; 2018.
- Jay M. R package generalhoslem [cited 2022 Jun 24]. https://cran.r-project.org/web/packages/generalhoslem
- Bursac Z, Gauss CH, Williams DK, Hosmer DW. Purposeful selection of variables in logistic regression. Source Code Biol Med. 2008;3:17. DOIPubMedGoogle Scholar
- Zhang Z. Model building strategy for logistic regression: purposeful selection. Ann Transl Med. 2016;4:111. DOIPubMedGoogle Scholar
- Naseer U, Løbersli I, Hindrum M, Bruvik T, Brandal LT. Virulence factors of Shiga toxin-producing Escherichia coli and the risk of developing haemolytic uraemic syndrome in Norway, 1992-2013. Eur J Clin Microbiol Infect Dis. 2017;36:1613–20. DOIPubMedGoogle Scholar
- Ylinen E, Salmenlinna S, Halkilahti J, Jahnukainen T, Korhonen L, Virkkala T, et al. Hemolytic uremic syndrome caused by Shiga toxin-producing Escherichia coli in children: incidence, risk factors, and clinical outcome. Pediatr Nephrol. 2020;35:1749–59. DOIPubMedGoogle Scholar
- Brandal LT, Wester AL, Lange H, Løbersli I, Lindstedt BA, Vold L, et al. Shiga toxin-producing escherichia coli infections in Norway, 1992-2012: characterization of isolates and identification of risk factors for haemolytic uremic syndrome. BMC Infect Dis. 2015;15:324. DOIPubMedGoogle Scholar
- Murphy V, Carroll AM, Forde K, Broni R, McNamara EB. Verocytotoxin Escherichia coli–associated haemolytic uraemic syndrome. Ir Med J. 2020;113:5.PubMedGoogle Scholar
- Bai X, Zhang J, Hua Y, Jernberg C, Xiong Y, French N, et al. Genomic insights into clinical Shiga toxin–producing Escherichia coli strains: a 15-year period survey in Jönköping, Sweden. Front Microbiol. 2021;12:
627861 . DOIPubMedGoogle Scholar - Public Health England. Interim public health operational guidance for Shiga toxin–producing Escherichia coli (STEC) including STEC (O157 and non-O157) infections. 2018 [cited 2021 Nov 1]. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/732569/Interim_public_health_operational_guidance_for_STEC_PDF.pdf
- Public Health England. Shiga toxin–producing Escherichia coli 2019 [cited 2021 Nov 1]. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/774291/STEC_O157_report.pdf
- Matussek A, Mernelius S, Chromek M, Zhang J, Frykman A, Hansson S, et al. Genome-wide association study of hemolytic uremic syndrome causing Shiga toxin-producing Escherichia coli from Sweden, 1994-2018. Eur J Clin Microbiol Infect Dis. 2023;42:771–9. DOIPubMedGoogle Scholar
- Koutsoumanis K, Allende A, Alvarez-Ordóñez A, Bover-Cid S, Chemaly M, Davies R, et al. Pathogenicity assessment of Shiga toxin–producing Escherichia coli (STEC) and the public health risk posed by contamination of food with STEC. EFSA J. 2020;18. DOIGoogle Scholar
- Hellinga HW, Evans PR. Nucleotide sequence and high-level expression of the major Escherichia coli phosphofructokinase. Eur J Biochem. 1985;149:363–73. DOIPubMedGoogle Scholar
- Zhang D, Li SH-J, King CG, Wingreen NS, Gitai Z, Li Z. Global and gene-specific translational regulation in Escherichia coli across different conditions. PLOS Comput Biol. 2022;18:
e1010641 . DOIPubMedGoogle Scholar - Grass G, Otto M, Fricke B, Haney CJ, Rensing C, Nies DH, et al. FieF (YiiP) from Escherichia coli mediates decreased cellular accumulation of iron and relieves iron stress. Arch Microbiol. 2005;183:9–18. DOIPubMedGoogle Scholar
- Ouyang A, Gasner KM, Neville SL, McDevitt CA, Frawley ER. MntP and YiiP contribute to manganese efflux in Salmonella enterica serovar Typhimurium under conditions of manganese overload and nitrosative stress. Microbiol Spectr. 2022;10:
e0131621 . DOIPubMedGoogle Scholar - Lee J, Hiibel SR, Reardon KF, Wood TK. Identification of stress-related proteins in Escherichia coli using the pollutant cis-dichloroethylene. J Appl Microbiol. 2010;108:2088–102.PubMedGoogle Scholar
- Fukushima K, Kumar SD, Suzuki S. YgiW homologous gene from Pseudomonas aeruginosa 25W is responsible for tributyltin resistance. J Gen Appl Microbiol. 2012;58:283–9. DOIPubMedGoogle Scholar
- Juárez-Rodríguez MD, Torres-Escobar A, Demuth DR. ygiW and qseBC are co-expressed in Aggregatibacter actinomycetemcomitans and regulate biofilm growth. Microbiology (Reading). 2013;159:989–1001. DOIPubMedGoogle Scholar
- Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol. 1999;32:1090–102. DOIPubMedGoogle Scholar
- Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833–5. DOIPubMedGoogle Scholar
- Lees JA, Galardini M, Bentley SD, Weiser JN, Corander J. pyseer: a comprehensive tool for microbial pangenome-wide association studies. Bioinformatics. 2018;34:4310–2. DOIPubMedGoogle Scholar
- Earle SG, Wu CH, Charlesworth J, Stoesser N, Gordon NC, Walker TM, et al. Identifying lineage effects when controlling for population structure improves power in bacterial association studies. Nat Microbiol. 2016;1:16041. DOIPubMedGoogle Scholar
- Farhat MR, Freschi L, Calderon R, Ioerger T, Snyder M, Meehan CJ, et al. GWAS for quantitative resistance phenotypes in Mycobacterium tuberculosis reveals resistance genes and regulatory regions. Nat Commun. 2019;10:2128. DOIPubMedGoogle Scholar
1These authors were co–principal investigators, contributed equally to this work, and share first authorship.