Volume 10, Number 11—November 2004
Research
Topographic Changes in SARS Coronavirus–infected Cells during Late Stages of Infection
Cite This Article
Citation for Media
Abstract
Scanning electron and atomic force microscopy was used for the first time to view the maturation of the severe acute respiratory syndrome–associated coronavirus at the cell surface. The surface form of the cells at advanced infection displayed prolific pseudopodia that, in addition to the rest of the plasma membrane, were also active sites of virus release. High magnification of the maturing virus particles showed a rosette appearance with short knoblike spikes under both the scanning electron and atomic force microscopes. The final expulsion step of the maturing virus particles seemed to result in some disruptions to the plasma membrane. The cytoskeletal network along the edge of the infected cells was enhanced and could be involved in transporting and expelling the progeny virus particles. Thickening of the actin filaments at the cell edge provided the bending force to extrude the virus particles.
A new human coronavirus was identified during the recent outbreak of severe acute respiratory syndrome (SARS) (1–4). The outbreak started during November 2002 in southern China and then spread to Hong Kong, Vietnam, Canada, and Singapore in early 2003. Sequence analyses of various isolates have indicated that the virus is genetically distinct from all known coronaviruses (5–7). Phylogenetic analysis suggests that the SARS-associated coronavirus (SARS-CoV) does not fit in the three currently known groups of coronaviruses (1,5,6,8), which suggests that this is a new virus, not a result of mutation or recombination of known coronaviruses.
Coronavirus infections are common in both domestic animals and humans (9). However, the known human coronaviruses often cause coldlike symptoms, whereas recent infections caused by SARS-CoV do not. The rate of death for SARS infections is 7%–10%, depending on the age of the patients (2).
SARS-CoV grows well in Vero E6 cells (1,2,10) and enters cells by direct fusion of the virus envelope with the plasma membrane (11). The fusion process involving the S glycoprotein is pH independent (12). Once internalized, the virus core uncoats, revealing flattened, disc-shaped, and electron-dense nucleocapsids described as “doughnut-shaped” (10,11). The uncoated nucleocapsids are found within large, smooth, double-membrane vacuoles together with membrane whorls (11). These membrane whorls are postulated to be replication complexes for the virus since they appear very early (within 30 minutes) after infection. Other reports have described double-membrane vesicles as sites of replication for coronavirus (Linder strain) (13), mouse hepatitis virus (14), and SARS virus (15). The latent period observed was 5–6 h postinfection (10). However, a short latent period is common among coronaviruses (16).
Coronavirus infections can be cytocidal for the cells; or, in some cases, persistent infection can result (17). The outcome of the infection is dependent on the virus strains and cell types. Unlike infection with the hepatitis C virus-229E, wherein virus production can continue for weeks without any expression of cytopathic effects (18,19), infection with SARS-CoV produces copious progeny virus particles within the first 12 h (10). The site of assembly of SARS-CoV was at the Golgi complexes, similar to previous reports for other coronaviruses (20–22). After assembly, the virus progeny particles are transported in vesicles to the cell periphery for release.
The aim of this study was to use scanning electron and atomic force microscopes to investigate changes in the surface topography of SARS-CoV–infected cells at late infection. The results can assist in further understanding how SARS CoV interacts with infected cells at late infection. Thus far, replication studies on SARS-CoV were performed with transmission electron microscopy, which showed detailed intracellular changes during replication in two dimensions. Both scanning electron and atomic force microscopy can provide holistic and three-dimensional views as infection progress.
Cells and Virus
SARS-CoV (2003VA2774) used for this study was isolated from a SARS patient in Singapore by the Department of Pathology, Singapore General Hospital. The virus was grown in Vero E6 cells (ATCC: C1008) in the Environmental Health Institute, National Environmental Agency, Singapore. Infection of the cells grown on coverslips and subsequent fixation (5% glutaraldehyde) of the infected cells at appropriate times were performed at that institute. The microscopy work on the fixed infected cells was performed at the Electron Microscopy Unit, National University of Singapore.
Scanning Electron Microscopy
Vero cells were grown to 70% confluency on sterile glass coverslips in 24-well tissue culture plates before infection with 100 μL of SARS-CoV for 1 h (multiplicity of infection = 10). Maintenance media supplemented with 2% fetal calf serum was added to the wells, and the infected cells were incubated in 37°C incubator with 5% carbon dioxide.
At an appropriate time after infection, the infected cells on the coverslips were fixed with 5% glutaraldehyde overnight. The coverslips were washed with phosphate-buffered saline before postfixation in 1% osmium tetroxide for 1 h. The coverslips were then washed with distilled water and dehydrated through a series of increasing concentration (25%–100%) of ethanol. Cells on the coverslips were further subjected to critical point drying for 1.5 h and left in a 37°C oven overnight. Subsequently, the cells on the coverslips were sputter coated with gold (thickness of 10 nm) and viewed under the XL30 Field Emission Gun scanning electron microscope (FEI Company, Enidhoven, the Netherlands) at 10 kV.
Atomic Force Microscopy
Infected cells were processed similarly. Normally, samples for the atomic force microscopy should be subjected to minimal processing so that the samples are close to their natural condition. However, in view of the pathogenicity of SARS-CoV, only fixed and gold-coated samples were used for this study. The NanoScope IV MultiMode atomic force microscope was used (Veeco, Woodbury, NY). Forced modulation etched silicon probes were used for imaging (dry tapping mode) infected cells. Hard tapping using appropriate amplitude setpoints was performed with some samples to show subsurface structures.
Negative Staining
Purified virus fixed in 2.5% glutaraldehyde was put onto a formvar carbon-coated grid and allowed to adsorb for a few minutes before being stained with 1% phosphotungstenic acid for 1 min. The excess fluid was blotted and the grid left to dry before viewing under CM120 BioTwin TEM (FEI Company, Enidhoven, the Netherlands).
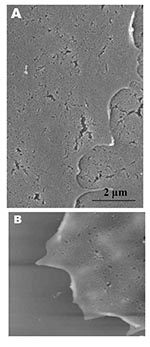
Both scanning electron and atomic force microscopy showed that the uninfected Vero cells were flat and without prominent surface morphology (Figure 1). Pseudopodia, where present, were not extensive (Figure 1A and 1B).
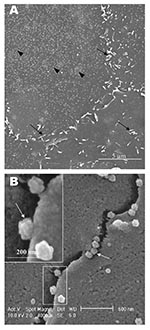
In the transmission electron microscopy studies (10,11,15), SARS-CoV replicated very rapidly and produced large amounts of virus after 6 h of infection. The scanning electron microscopy confirmed that, for some infected cells (15 h postinfection), a large quantity of extracellular virus was present (Figure 2A, arrowheads) on the whole cell surface. However, very few virus particles were on the neighboring cell (top right), indicating a nonsynchronous infection. The scanning electron microscopy images showed a holistic view of SARS-CoV–infected cells compared to ultrathin sections in transmission electron microscopy. Another virus-induced change clearly demonstrated by using the scanning electron microscopy was the proliferation of pseudopodia on the infected cells and in particular, at the edge of these cells (Figure 2A, arrows compared to Figure 1).
At higher magnification, progeny virus particles protruded at the cell periphery (Figure 2B, arrow). In the inset (boxed area), a virus particle was seen in the process of extrusion (arrow) after the fusion of the transport vesicle and the plasma membrane. The knoblike spikes surrounding the coronavirus were clearly visible (Figure 2C, arrows). SARS-CoV spikes appeared short and stubby (16–17 nm) when compared to those of other coronaviruses (20 nm). This feature gave the virus a rosettelike appearance when viewed under the scanning electron microscope (arrowheads indicate extruded virus particles). The average size of the extracellular virus particles was 100–130 nm. The gold sputter coating can also increase in the diameters of the virus particles.
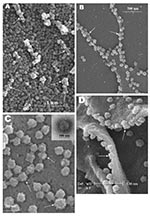
From 15 to 24 h after infection, the virus was exported prolifically at the pseudopodia and cell surfaces (Figure 3A–C, arrows). The surface imaging clearly showed the profuse presence of extracellular virus (arrows). High magnification scanning electron microscopy images of the SARS-CoV form and structure (Figure 3C, arrows) appeared to correlate well with those images that used negative staining and TEM (Figure 3C, inset). The knoblike spikes were short and stubby in the negative staining image as well. Online Figure 3D (available at http://www.cdc.gov/ncidod/EID/vol10no11/04-0195-G3D.htm) shows virus particles were also exported out from the surface of the pseudopodia (arrows).
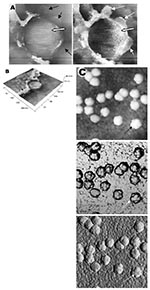
A virus particle in the process of extrusion at the cell plasma membrane was captured with the atomic force microscope at 15 hours after infection. Although the proposed mechanism for export of the virus to the extracellular space is through fusion of the transport vesicle membrane at the cell surface, this process seemed to result in localized breaching at the plasma membrane, where the virus extrusion occurred (Figure 4A, thin arrows). Although fixed and gold-coated samples were used in this study, the atomic force microscope delivered high-resolution images. Unfortunately, the knoblike spikes for this virus were not well illustrated in Figure 4A. A three-dimensional reconstruction (Figure 4B) shows that the virus particle was extruding from a much-thickened cell periphery (arrows). The knoblike structures on the virus surface were further confirmed by atomic force microscopy (online Figure 4C, available at http://www.cdc.gov/ncidod/EID/vol10no11/04-0195-G4C.htm).
The thickened edges of the infected cells were ruffled and appeared to comprise layers of folded membranes (Figure 5A and 5B and online Figure 5C, available at http://www.cdc.gov/ncidod/EID/vol10no11/04-0195-G5C.htm). The layered/folded effects at the edge of cells were pronounced in the height image under the atomic force microscopy and scanning electron micrographs. The arrowheads show the virus particles.
Virus particles could still be exported out of the puffy edge (Figure 6A, arrows). A three-dimensional reconstruction (Figure 6B) of the height image in Figure 6A shows puffy fronts of the cell edge (arrows) with many virus particles just underneath the surface awaiting extrusion. The large number of progeny virus particles at the cell edge may have resulted in this thickened appearance. Virus particles (arrowheads) were present on other parts of the cell surface as well.
Closer examination of the virus-induced changes at the subcellular surfaces of the infected cells, by using the hard tapping mode under the atomic force microscope, showed the involvement of the cell cytoskeleton at late infection. In Figure 7A, gross thickening of the cell skeletal filaments was seen in the cytoplasm (arrowhead) and pseudopodia (arrows). At higher resolution, thickening of the filaments at the edge of cells was obvious (Figure 7B, arrows). These filaments, which ran parallel to the cell edge, could be the enhanced actin filaments, and together with the accumulated progeny virus particles, could have caused the bulky, puffy-cell periphery.
By using transmission electron microscopy, recent studies (10,11) showed the entry events and prolific growth of SARS-CoV in Vero E6 cells. SARS-CoV enters the cell by direct fusion and has a latent period of only 6 h. High numbers of progeny virus particles assemble in the swollen Golgi sacs before export to the external surface.
Transmission electron microscopy of ultrathin sections gave good intracellular information but was not able to give a gross morphologic landscape of the infected cells. Surface topographic changes induced by SARS-CoV at maturation and late stages of infections were the focus of this study. The virus-induced modifications at the cell surface or subcellular surface could relate to the eventual destruction of the infected cells as well as shed light on the extrusion mechanism of the progeny virus particles from the cell surface.
Scanning electron microscopy, an established technique, gives a three-dimensional overview of the virus and the infected cell surfaces. Another high-resolution technology used in this study is the atomic force microscope, which gives atomic resolution. It is also gaining popularity in areas of life science research (23–29). Most of these studies were on purified macromolecules. However, the atomic force microscopy has also gained a foothold in virology in recent years (30–33). A recent study on HIV and HIV-infected lymphocytes (34) demonstrated the strength of this technique for virology.
The application of these two selected techniques to study the late SARS virus–induced changes in Vero cells was rewarding. The SARS-CoV knobby/rosettelike structures were seen in a three-dimensional form under the scanning electron and atomic microscopy (Figures 2B, 2C, 4A). The spikes seemed shorter (16–17 nm) than those of other coronaviruses. At this stage, it is speculative if this could be due to the lack of the hemagglutinin-esterase protein (8,35) in the spike glycoprotein of this virus. Further structural and functional studies should be performed to investigate this aspect and its relation to virus virulence.
The scanning electron microscopy studies showed prolific SARS-CoV on infected cell surface 15 hours after infection. Unlike ultrathin sectioning in transmission electron microscopy, the scanning techniques allow cell and virus surfaces to be viewed without invasive manipulation. In addition to the large amount of extracellular virus particles on most cells, proliferation of the pseudopodia in the infected cells was pronounced (Figure 2A compared to Figure 1). These pseudopodia increase the surface area of the cells as active maturation sites of virus (Figures 3A and 3B).
Although the scanning electron microscope was able to show virus particles in the process of extruding (Figure 2B, Figure 3A and B) from the cells, the image derived with the atomic force microscope was superior in resolution. A virus particle was seen pushing out of the cell plasma membrane (Figure 4A), which resulted in localized loss of membrane integrity at the site. Since prolific extrusion of the progeny virus particles occurred at this late stage of infection, the frequent loss of plasma membrane integrity could compromise the physiologic status of the infected cells and lead to cell death.
Fifteen h after infection, ruffled, puffy infected cell peripheries were demonstrated (Figures 4B, 5, and 6) and not seen in uninfected cells (Figure 1). This feature was not obvious under the transmission electron microscopy (11). Subcellular imaging of the thickened edge of the cells showed numerous progeny virus particles awaiting extrusion (Figure 6B, arrows). The actin filaments that were parallel to the cell edge appeared to have thickened (Figures 7A and B compared to Figures 1A and lB). The enhanced presence of the actin filaments could assist in providing the bending force to expel the progeny virus particles to the exterior. Bohn and colleagues (36) suggested that the forces resulting from the vectorial growth of the actin filaments contributed to membrane bending at the site of virus maturation. Actin filaments have also been reported to be directly involved in the budding of both enveloped DNA and RNA viruses (37–40).
In summary, the cellular cytoskeleton network is involved in the SARS-CoV maturation and possibly replication process. The constant loss of membrane integrity attributable to the prolific progeny virus extrusion resulted in disintegration of infected cells.
Dr. Ng is an associate professor at the Department of Microbiology, National University of Singapore. Her research interests are virology (main focus is on flaviviruses) and microscopy techniques.
Acknowledgments
Purified virus fixed in 2.5% glutaraldehyde was provided by the Environmental Health Institute.
The microscopy work was supported by the Academic Research Fund, National University of Singapore (Grant no. R-182-000-055-112).
References
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–76. DOIPubMedGoogle Scholar
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–66. DOIPubMedGoogle Scholar
- Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM, A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–94. DOIPubMedGoogle Scholar
- Poutanen SM, Low DE, Henry B, Finkelstein S, Rose D, Green K, Identification of severe acute respiratory syndrome in Canada. N Engl J Med. 2003;348:1995–2005. DOIPubMedGoogle Scholar
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, C haracterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–9. DOIPubMedGoogle Scholar
- Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–404. DOIPubMedGoogle Scholar
- Ruan YJ, Wei CL, Ling AE, Vega VB, Thoreau H, Se Thoe SY, Comparative full length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–85. DOIPubMedGoogle Scholar
- Lai MMC, Holmes KV. Coronaviridae and their replication. In: Fields BM, Knipe DM, Howley PM, Chanock RM, Melnick JL, Monath TP, et al. Field’s virology, 3rd ed. Philadelphia: Lippincott-Raven; 2001. p. 1163–85.
- Ng ML, Tan SH, See EE, Ooi EE, Ling AE. Prolific replication of SARS coronavirus in E6 cells. J Gen Virol. 2003;84:3291–303. DOIPubMedGoogle Scholar
- Ng ML, Tan SH, See EE, Ooi EE, Ling AE. Entry and early events of severe acute respiratory syndrome coronavirus. J Med Virol. 2003;71:323–31. DOIPubMedGoogle Scholar
- Xiao X, Chakraborti S, Dimitrov AS, Gramatikoff K, Dimitrov DS. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem Biophys Res Commun. 2003;312:1159–64. DOIPubMedGoogle Scholar
- Oshiro LS, Schieble JH, Lennette EH. Electron microscopic studies of coronavirus. J Gen Virol. 1971;12:161–8. DOIPubMedGoogle Scholar
- Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol. 2002;76:3697–708. DOIPubMedGoogle Scholar
- Goldsmith CS, Tatti KM, Ksiazek TG, Rollin PE, Comer JA, Lee WW, Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis. 2004;10:320–6.PubMedGoogle Scholar
- Sturman LS, Takemoto KK. Enhanced growth of a murine coronavirus in transformed mouse cells. Infect Immun. 1972;6:501–7.PubMedGoogle Scholar
- Wege H, Siddell S, ter Meulen V. The biology and pathogenesis of coronaviruses. Curr Top Microbiol Immunol. 1982;99:165–200.PubMedGoogle Scholar
- Chaloner-Larsson G, Johnson-Lussenburg CM. Establishment and maintenance of a persistent infection of L123 cells by human coronavirus strain 299E. Arch Virol. 1981;69:117–29. DOIPubMedGoogle Scholar
- Holmes KV, Behnke JN. Evolution of a coronavirus during persistent infection in vitro. Adv Exp Med Biol. 1981;142:287–99.PubMedGoogle Scholar
- Wilhelmsen KC, Leibowitz JL, Bond CW, Robb JA. The replication of murine coronaviruses in enucleated cells. Virology. 1981;110:225–30. DOIPubMedGoogle Scholar
- Tooze J, Tooze SA. Infection of AtT20 murine pituitary tumor cells by mouse hepatitis virus strain A59: virus budding is restricted to the Golgi region. Eur J Cell Biol. 1985;37:203–12.PubMedGoogle Scholar
- Tooze J, Tooze S, Warren G. Replication of coronavirus MHV-A59 in sac-cells: determination of the first site of budding of progeny virions. Eur J Cell Biol. 1984;33:281–93.PubMedGoogle Scholar
- Allen MJ, Balooch M, Subbiah S, Tench RJ, Siekhaus W, Balhorn R. Scanning tunneling microscope images of adenine and thymine at atomic resolution. Scanning Microsc. 1991;5:625–30.PubMedGoogle Scholar
- Almqvist N, Backman L, Fredriksson S. Imaging human erythrocyte spectrin with atomic force microscopy. Micron. 1994;5:227–32. DOIPubMedGoogle Scholar
- Baranauskas V, Vidal BC, Parizotto NA. Observation of geometric structure of collagen molecules by atomic force microscopy. Appl Biochem Biotechnol. 1998;69:91–7. DOIPubMedGoogle Scholar
- Baldwin PM, Davies MC, Melia CD. Starch granule surface imaging using low-voltage scanning electron microscopy and atomic force microscopy. Int J Biol Macromol. 1998;21:103–7. DOIPubMedGoogle Scholar
- Deleu M, Nott K, Brasseur R, Jacques P, Thonart P, Dufrene YF. Imaging mixed lipid monolayers by dynamic atomic force microscopy. Biochim Biophys Acta. 2001;1513:55–62. DOIPubMedGoogle Scholar
- Beckmann M, Kolb HA, Lang F. Atomic force microscopy of biological cell membranes: from cells to molecules. Microscopy and Analysis. 1995;Jan:7–9.
- Chen CH, Hansma HG. Basement membrane macromolecules: insights from atomic force microscopy. J Struct Biol. 2000;131:44–55. DOIPubMedGoogle Scholar
- Falvo MR, Washburn S, Superfine R, Finch M, Brooks FP Jr, Chi V, Manipulation of individual viruses: friction and mechanical properties. Biophys J. 1997;72:1396–403. DOIPubMedGoogle Scholar
- Ohnesorge FM, Horber JK, Haberle W, Czerny CP, Smith DP, Binnig G. AFM review study on poxviruses and living cells. Biophys J. 1997;73:2183–94. DOIPubMedGoogle Scholar
- Drygin YF, Bordunova OA, Gallyamov MO, Yaminsky IV. Atomic force microscopy examination of tobacco mosaic virus and virion RNA. FEBS Lett. 1998;425:217–21. DOIPubMedGoogle Scholar
- Kiselyova OI, Yaminsky IV, Karger EM, Frolova OY, Dorokhov YL, Atabekov JG. Visualization by atomic force microscopy of tobacco mosaic virus movement protein-RNA complexes formed in vitro. J Gen Virol. 2001;82:1503–8.PubMedGoogle Scholar
- Kuznetsov YG, Victoria JG, Robinson WE Jr, McPherson A. Atomic force microscopy investigation of human immunodeficiency virus (HIV) and HIV-infected lymphocytes. J Virol. 2003;77:11896–909. DOIPubMedGoogle Scholar
- Holmes KV, Enjuanes L. The SARS coronavirus: a postgenomic era. Science. 2003;300:1377–8. DOIPubMedGoogle Scholar
- Bohn W, Rutter G, Hohenberg H, Mannweiler K, Nobis P. Involvement of actin filaments in budding of measles virus: studies on cytoskeletons of infected cells. Virology. 1986;149:91–106. DOIPubMedGoogle Scholar
- Lanier IM, Volkman LE. Actin binding and nucleation by Autographa California M nucleopolyhedrovirus. Virology. 1998;243:167–77. DOIPubMedGoogle Scholar
- Ravkov EV, Nichol ST, Peters CL, Compans RW. Role of actin microfilaments in Black Creek Canal virus morphogenesis. J Virol. 1998;72:2865–70.PubMedGoogle Scholar
- Boulanger D, Smith T, Skinner MA. Morphogenesis and release of fowlpox virus. J Gen Virol. 2000;81:675–87.PubMedGoogle Scholar
- Chu JJH, Choo BGH, Lee JWM, Ng ML. Actin filaments participate in West Nile (Sarafend) virus maturation process. J Med Virol. 2003;71:463–72. DOIPubMedGoogle Scholar
Figures
Cite This ArticleTable of Contents – Volume 10, Number 11—November 2004
| EID Search Options |
|---|
|
|
|
|
|
|



Please use the form below to submit correspondence to the authors or contact them at the following address:
Mah-Lee Ng, Department of Microbiology, 5 Science Drive 2, National University of Singapore, Singapore 117597; fax: 65-6776-6872
Top