Volume 21, Number 1—January 2015
Research
Protocol for Metagenomic Virus Detection in Clinical Specimens1
Figure 10
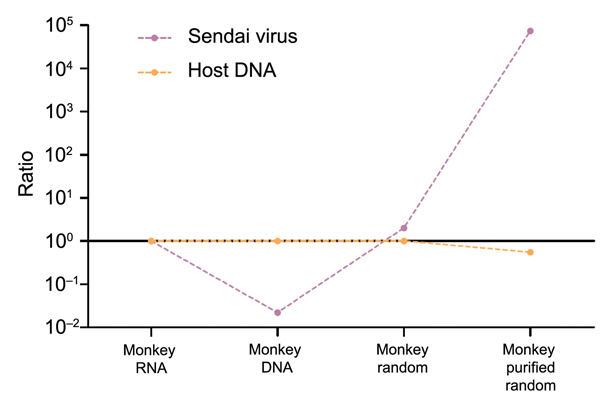
Figure 10. Changes in virus-to-host nucleic acid signal-to-noise ratio during development of tissue-based universal virus detection for viral metagenomics (TUViD-VM) protocol. Next-generation sequencing results for virus-infected marmoset tissue comparatively sequenced were obtained by using 4 approaches: standard RNA library preparation (Monkey RNA), standard DNA library preparation (Monkey DNA), DNA library from random-amplified marmoset tissue (Monkey random), and DNA library from random-amplified marmoset tissue prepared by using TUViD-VM (Monkey purified random). The y-axis is log-scaled.
1Preliminary results of this study were presented at the Biodefense and Emerging Infectious Diseases Meeting, January 29, 2014, Washington DC, USA.
Page created: January 05, 2015
Page updated: January 05, 2015
Page reviewed: January 05, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.