Volume 26, Number 12—December 2020
Dispatch
Sensitive Detection of SARS-CoV-2–Specific Antibodies in Dried Blood Spot Samples
Cite This Article
Citation for Media
Abstract
Dried blood spot (DBS) samples can be used for the detection of severe acute respiratory syndrome coronavirus 2 spike antibodies. DBS sampling is comparable to matched serum samples with a relative 98.1% sensitivity and 100% specificity. Thus, DBS sampling offers an alternative for population-wide serologic testing in the coronavirus pandemic.
A confirmed diagnosis of acute coronavirus disease (COVID-19) depends on the detection of RNA from the causative pathogen, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In contrast, although serologic testing is less useful for diagnosing the acute stages of infection, it can aid in diagnosing atypical manifestations of SARS-CoV-2 infection (M. Perez-Toledo et al., unpub. data, https://doi.org/10.1101/2020.06.05.20123117) and in determining prior virus exposure at a population level (1), knowledge which could substantially influence public health and social policies (2,3).
Currently, antibody testing for SARS-CoV-2 uses serum or plasma collected by venipuncture. The use of such sampling in large-scale seroepidemiologic studies is limited by logistic challenges, resources, and costs, as well as the risk for SARS-CoV-2 exposure from direct patient contact. In contrast, dried blood spot (DBS) sampling is simple, inexpensive, and can be self-collected and then sent by postal services to laboratories for processing (4). It is a well-established method for detecting antibodies against various infections (5,6), and antibodies collected by DBS are stable for prolonged periods (7). Moreover, DBS sampling provides a solution to widening access to serologic platforms in low- and middle-income countries. Nevertheless, the potential role of DBS sampling in studying SARS-CoV-2 seroprevalence has not been fully explored, and knowledge regarding the recovery of antibody from the DBS is limited. We describe the validation of DBS samples against matched serum in a highly sensitive and specific SARS-CoV-2 ELISA.
We collected 87 samples from 80 volunteers at the University Hospitals Birmingham NHS Foundation Trust (under approved protocol for blood donations use in clinical assays, UK Research Ethics Committee reference no. 2002/201 and Clinical Immunology Service Reference no. ERN_16-178) during May 18–June 3, 2020. Three matched samples were from SARS-CoV-2 serum antibody–negative volunteers. The remaining samples were from SARS-CoV-2 serum antibody–unknown volunteers; 5 volunteers provided duplicate and 1 volunteer provided triplicate matched samples (Appendix Figure). To refine negative thresholds, we included 17 pre–August 2019 DBS-only samples (UK Research Ethics Committee reference no. 2002/20, Integrated Research Application System reference no. 132132, University Hospitals Birmingham project reference no. RRK4136). Volunteers were healthy at the time of sampling. Thirty-one matched samples (31/87 [35.6%]) were from PCR-positive volunteers, on average, 54 days (SD + 17 days) from reported symptom onset and 45 days (SD + 15 days) from PCR testing. All participants were anonymized, and SARS-CoV-2 PCR status was recorded as positive or unknown.
For DBS collection, we collected capillary blood samples onto forensic-grade 226 DBS cards (Ahlstrom Munksjo, https://www.ahlstrom-munksjo.com) by using finger-prick lancets (4,8). We stored DBS cards at room temperature in individual sample bags with desiccant. Concomitantly, we collected venous blood from volunteers and separated serum by using centrifugation at 9,700 × g for 5 min at room temperature. Laboratory analysis was blinded to PCR status, and we reported SARS-CoV-2–specific antibody results as positive, negative, or equivocal.
To elute antibody from DBS cards, we isolated individual preperforated DBS spots by using a sterile pipette tip and placed them into a universal tube at a ratio of 1 spot to 250 µL 0.05% phosphate-buffered saline (PBS)–Tween 20 (PBS-T) (PBS, Oxoid; Tween-20; Sigma-Aldrich, https://www.sigmaaldrich.com). We briefly vortexed and incubated tubes overnight at room temperature. We then harvested DBS eluate into a microtube and centrifuged it at 10,600 × g for 10 min at room temperature. We stored eluate at 4°C for <14 days in accordance with standard protocols (4). We quantified total IgG, IgA, and IgM concentrations in matched serum and DBS eluate, plus pre–August 2019 DBS samples, with nephelometry by using the automated COBAS 6000 (Roche, https://www.roche.com).
We performed a highly sensitive and specific in-house ELISA (now under peer review) to measure IgG, IgA and IgM against soluble, stabilized, trimeric SARS-CoV-2 spike (S) glycoprotein (9,10), as previously described (S.E. Faustini et al., unpub. data, https://doi.org/10.1101/2020.06.16.20133025). In brief, we coated Nunc 96-well plates (ThermoFisher, https://www.thermofisher.com) with 50 µL of 2 µg/mL S glycoprotein (M. Perez-Toledo et al.; S.E. Faustini et al.). We blocked plates and diluted samples with 2% BSA 0.1% PBS-T (PBS, Oxoid; Tween-20 and BSA, Sigma-Aldrich) at starting dilutions of 1:3 DBS eluate and 1:15 serum, with 3-fold serial dilutions; or single dilutions of 1:10 DBS eluate and 1:100 serum. We diluted mouse monoclonal anti–human horseradish peroxidase conjugated antibodies (anti–IgG R-10 1:8,000, anti–IgA MG4.156 1:4,000, and anti–IgM AF6 1:2,000; Abingdon Health, https://www.abingdonhealth.com) in 0.1% PBS-T. We developed plates with TMB Core (Bio-Rad, https://www.bio-rad.com) and stopped them after 5 min with 0.2M H2SO4 (Sigma-Aldrich). We recorded optical densities at 450 nm (OD450) by using the Dynex Revelation (Dynex Technologies, https://www.dynextechnologies.com). We reported results as SARS-CoV-2 S antibody positive, negative, or equivocal. The cutoff for negativity was less than the highest negative control (DBS 0.399 OD450 and serum 0.449 OD450), and for positivity, the mean of the negative controls +3 SD (DBS 0.444 OD450 and serum 0.62 OD450); a result between this range was considered equivocal.
We performed statistical analyses by using Prism 8 (GraphPad, https://www.graphpad.com) and assessed correlations between continuous data by using Spearman’s rank test (p<0.05 was considered statistically significant). We assessed DBS sample ELISA performance, relative to the serum assay, by calculating the comparative sensitivity, specificity, and positive and negative predictive values, with 95 % CIs. We assessed the agreement between DBS and serum ELISA results by determining the Cohen κ coefficient and Bland-Altman mean-difference.
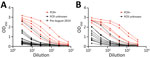
We performed quantification of total immunoglobulin concentrations in serum and DBS eluate. We observed 7- to 11-fold reduction in mean immunoglobulin concentration (IgG, IgA, and IgM) in DBS eluate compared with matched serum (Table 1). Matched serum and DBS titration curves showed the detection of SARS-CoV-2 S glycoprotein antibodies in both serum and DBS eluate with the limits of detection and the optimal detection dilution indicated (1:10 for DBS eluate and 1:100 for serum). PCR-positive matched samples showed higher responses, whereas pre–August 2019 DBS samples were negative across all dilutions (Figure 1).
We measured OD450 detected by ELISA for matched DBS eluate (diluted 1:10) and serum (diluted 1:100). We observed a significant correlation between matched serum and DBS samples (r = 0.96 [95% CI 0.93–0.97]; p<0.0001) (Figure 2, panel A) and minimal differences in results observed by sample type (Bland-Altman bias 0.11 + 0.20) (Figure 2, panel B). Discordance occurred between only 1 matched sample (κ = 0.975). Relative to serum samples, DBS samples achieved 98.11% sensitivity and 100% specificity for detecting S glycoprotein antibodies (Table 2); 100% of the PCR-positive samples (n = 31) were also antibody-positive in DBS eluate.
We show that DBS samples can be used for the detection of SARS-CoV-2–specific antibodies with results comparable to serum samples, supporting the findings of recent preliminary studies (11,12). Although individual laboratories should optimize DBS-derived antibody detection, considering dilution-factor and cutoff thresholds for their relevant downstream assay, these results demonstrate that DBS sampling could complement venipuncture for serologic assessments, such as seroprevalence studies, during the COVID-19 pandemic.
A current limitation of antibody assays is the necessity for venipuncture by skilled phlebotomists; DBS sampling overcomes this limitation and introduces the opportunity for wider population-level testing and improved surveillance in groups at heightened risk for infection. For example, DBS could be delivered using postal services (4) to patients with chronic conditions, the immunocompromised, and the elderly, all of which are groups disproportionately affected by COVID-19 (13). Furthermore, the DBS method is simple and inexpensive (4), which could enhance sampling in low- and middle-income countries, among groups where venipuncture is culturally unacceptable or in a geographically dispersed population.
Dr. Morley is a clinician specializing in public health, currently undertaking her PhD research, which is focused on humoral immunology, at the University of Birmingham. She worked on SARS-CoV-2 diagnostics and research during the COVID-19 pandemic at the University of Birmingham.
Acknowledgments
We would like to thank the University of Birmingham Clinical Immunology Service for their invaluable support in sample collection and processing. We also thank Cynthia D’Aguilar and Julie Williams for logistic support in sample collection. We are grateful for the expertise of Margaret Goodall in generating the mouse monoclonal anti–human horseradish peroxidase conjugated antibodies.
This work was supported by the Wellcome Trust and the National Institute for Health Research Birmingham Biomedical Research Centre at the University Hospitals Birmingham NHS Foundation Trust and the University of Birmingham. The views expressed are those of the authors and not necessarily those of the National Institute for Health Research or the Department of Health and Social Care. This project was supported by the Saving Lives Charity (UK Charity Commission no. 1144855) who kindly provided the dried blood spot collection cards. The work, conducted in Max Crispin’s laboratory, was funded by the International AIDS Vaccine Initiative, Bill and Melinda Gates Foundation through the Collaboration for AIDS Vaccine Discovery (grants nos. OPP1196345/INV-008813, OPP1084519, and OPP1115782), the Scripps Consortium for HIV Vaccine Development (National Institutes of Health National Institute for Allergy and Infectious Diseases grant no. AI144462), and the University of Southampton Coronavirus Response Fund.
S.T. is the medical director of the Saving Lives Charity. M.T.D. and M.G. report stocks in Abingdon Health (outside the submitted work).
References
- Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–4.PubMedGoogle Scholar
- Long Q-X, Tang X-J, Shi Q-L, Li Q, Deng H-J, Yuan J, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26:1200–4. DOIPubMedGoogle Scholar
- Shields A, Faustini SE, Perez-Toledo M, Jossi S, Aldera E, Allen JD, et al. SARS-CoV-2 seroprevalence and asymptomatic viral carriage in healthcare workers: a cross-sectional study. Thorax. 2020;
thoraxjnl-2020-215414 ; Epub ahead of print. DOIPubMedGoogle Scholar - Page M, Atabani SF, Wood M, Smit E, Wilson S, Atherton C, et al. Dried blood spot and mini-tube blood sample collection kits for postal HIV testing services: a comparative review of successes in a real-world setting. Sex Transm Infect. 2019;95:43–5. DOIPubMedGoogle Scholar
- Vázquez-Morón S, Ryan P, Ardizone-Jiménez B, Martín D, Troya J, Cuevas G, et al. Evaluation of dried blood spot samples for screening of hepatitis C and human immunodeficiency virus in a real-world setting. Sci Rep. 2018;8:1858. DOIPubMedGoogle Scholar
- Condorelli F, Scalia G, Stivala A, Gallo R, Marino A, Battaglini CM, et al. Detection of immunoglobulin G to measles virus, rubella virus, and mumps virus in serum samples and in microquantities of whole blood dried on filter paper. J Virol Methods. 1994;49:25–36. DOIPubMedGoogle Scholar
- Behets F, Kashamuka M, Pappaioanou M, Green TA, Ryder RW, Batter V, et al. Stability of human immunodeficiency virus type 1 antibodies in whole blood dried on filter paper and stored under various tropical conditions in Kinshasa, Zaire. J Clin Microbiol. 1992;30:1179–82. DOIPubMedGoogle Scholar
- Grüner N, Stambouli O, Ross RS. Dried blood spots—preparing and processing for use in immunoassays and in molecular techniques. J Vis Exp. 2015;97:52619. DOIPubMedGoogle Scholar
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–3. DOIPubMedGoogle Scholar
- Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–3. DOIPubMedGoogle Scholar
- McDade TW, McNally EM, Zelikovich AS, D’Aquila R, Mustanski B, Miller A, et al. High seroprevalence for SARS-CoV-2 among household members of essential workers detected using a dried blood spot assay. PLoS One. 2020;15:
e0237833 . DOIPubMedGoogle Scholar - Thevis M, Knoop A, Schaefer MS, Dufaux B, Schrader Y, Thomas A, et al. Can dried blood spots (DBS) contribute to conducting comprehensive SARS-CoV-2 antibody tests? Drug Test Anal. 2020;12:994–7. DOIPubMedGoogle Scholar
- Jordan RE, Adab P, Cheng KK. Covid-19: risk factors for severe disease and death. BMJ. 2020;368:m1198. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: September 24, 2020
1These senior authors contributed equally to this article.
Table of Contents – Volume 26, Number 12—December 2020
| EID Search Options |
|---|
|
|
|
|
|
|
![Effectiveness of DBS sampling for SARS-CoV-2 anti-spike glycoprotein detection. A) Correlation between matched DBS eluate (1:10) and serum (1:100) OD450 ELISA results (n = 87). Red circles indicate PCR-positive samples (n = 31). Black circles indicate PCR-unknown samples (n = 56). B) Bland-Altman mean-difference comparison of DBS eluate (1:10) and serum (1:100) OD450 ELISA results (dashed lines indicate 95% limits of agreement [−0.281 to 0.504]). DBS, dried blood spot; OD450, optical density at 450 nm; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.](/eid/images/20-3309-F2-tn.jpg)
Please use the form below to submit correspondence to the authors or contact them at the following address:
Matthew K. O’Shea. Institute of Immunology and Immunotherapy, College of Medical and Dental Sciences, University of Birmingham, Birmingham B15 2TT, UK
Top