Volume 29, Number 4—April 2023
Research
Adeno-Associated Virus 2 and Human Adenovirus F41 in Wastewater during Outbreak of Severe Acute Hepatitis in Children, Ireland
Figure 4
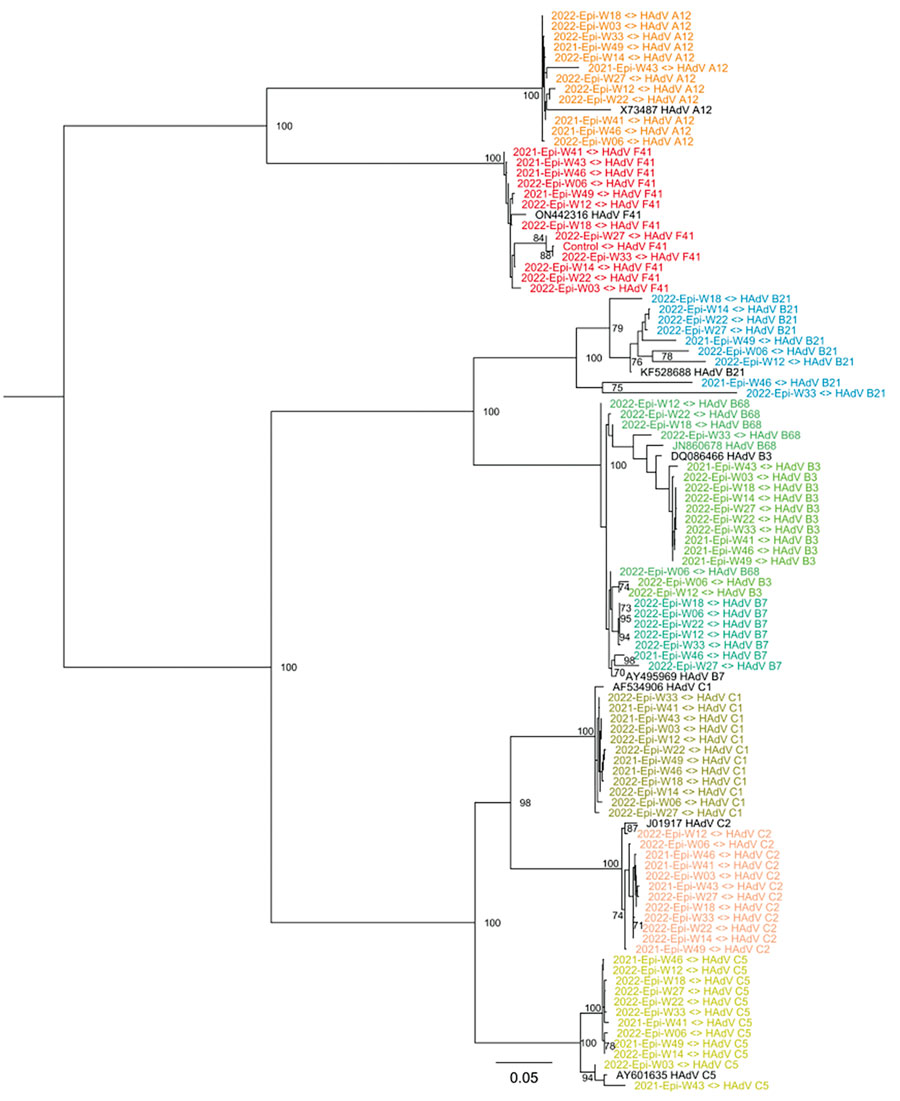
Figure 4. Maximum-likelihood phylogenetic tree of HAdV genetic diversity in wastewater samples collected during SAHUE outbreak in children, Ireland. The tree was inferred by using RAxML (https://github.com/stamatak/standard-RAxML). Branch support was estimated by using the bootstrap method with 100 repetitions and is shown next to the branches that have >70% support. Black text indicates reference sequences, identified by GenBank accession number. Colors indicate HAdV species and types. Scale bar indicates nucleotide substitutions per site. Epi-W, epidemiologic week; HAdV, human adenovirus; SAHUE, severe acute hepatitis of unknown etiology.
Page created: January 24, 2023
Page updated: March 20, 2023
Page reviewed: March 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.