Volume 30, Number 8—August 2024
Synopsis
Outbreak of Intermediate Species Leptospira venezuelensis Spread by Rodents to Cows and Humans in L. interrogans–Endemic Region, Venezuela
Figure 3
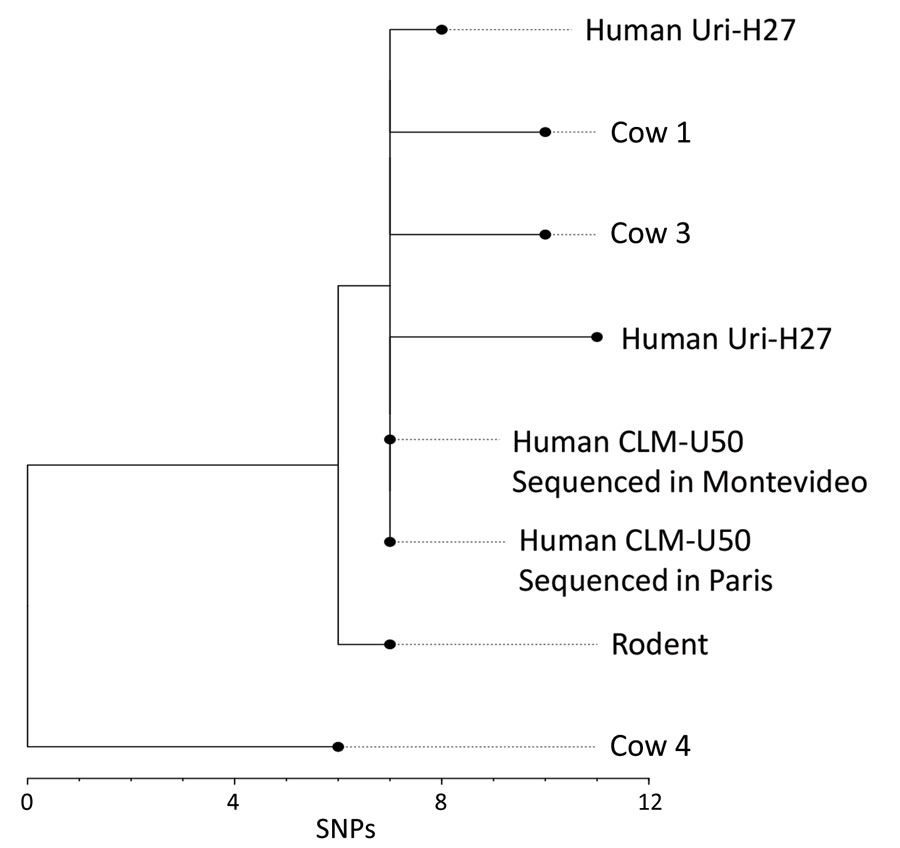
Figure 3. Phylogenetic analysis of Leptospira venezuelensis isolates in study of outbreak of intermediate species L. venezuelensis spread by rodents to cows and humans in L. interrogans–endemic region, Venezuela. Branch length indicates the number of SNPs separating L. venezuelensis strains. Phylogenetic tree was reconstructed according to comparisons of whole-genome sequences from 6 L. venezuelensis strains isolated from hospitalized leptospirosis patients in La Guaira State on the Caribbean coast of Venezuela, from rodents captured near the residences of hospitalized leptospirosis patients, and from dairy cows on a farm 30 km away from La Guaira State. Human isolate CLM-50 was sequenced at both the Institute Pasteur in Paris, France, and the Institute Pasteur in Montevideo, Uruguay. Human isolate Uri-H27 was sequenced twice at the Institute Pasteur in Paris; the genome of the isolate after many passages in culture contained 3 SNPs that were not present in the same isolate from an earlier passage. Scale bar indicates number of SNPs per site. SNP, single-nucleotide polymorphism.
1Current affiliation: Universidad de las Fuerzas Armadas ESPE, Santo Domingo, Ecuador.
2Current affiliation: Universidad de Buenos Aires, Buenos Aires, Argentina.
3Current affiliation: Institut Pasteur de Montevideo, Montevideo, Uruguay.
4Current affiliation: The University of North Carolina, Chapel Hill, North Carolina, USA.